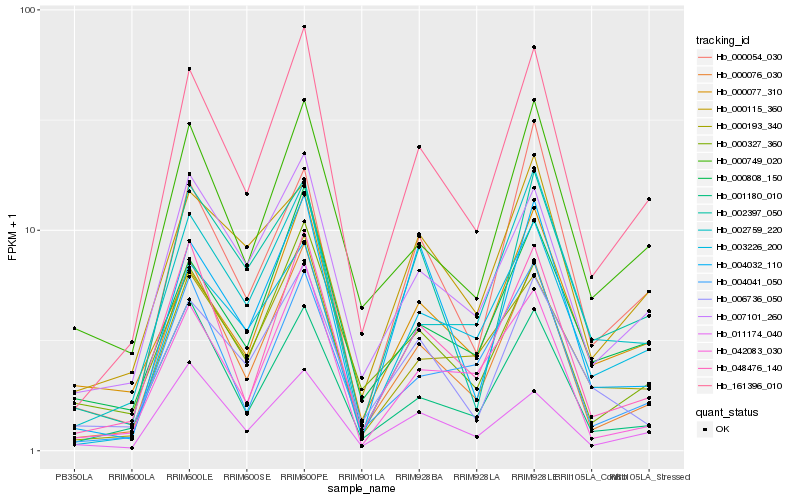
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_161396_010 |
0.0 |
- |
- |
hypothetical protein F383_26394 [Gossypium arboreum] |
| 2 |
Hb_000076_030 |
0.085687369 |
- |
- |
PREDICTED: microtubule-associated protein TORTIFOLIA1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_007101_260 |
0.0994640886 |
- |
- |
altered response to gravity (arg1), plant, putative [Ricinus communis] |
| 4 |
Hb_000193_340 |
0.1031507079 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000808_150 |
0.1110409578 |
- |
- |
PREDICTED: uncharacterized protein LOC105647444 [Jatropha curcas] |
| 6 |
Hb_048476_140 |
0.1124876548 |
- |
- |
PREDICTED: dynamin-related protein 1C [Jatropha curcas] |
| 7 |
Hb_002759_220 |
0.1154383067 |
- |
- |
altered response to gravity (arg1), plant, putative [Ricinus communis] |
| 8 |
Hb_000749_020 |
0.1159220095 |
- |
- |
Nucleotide-diphospho-sugar transferases superfamily protein isoform 1 [Theobroma cacao] |
| 9 |
Hb_003226_200 |
0.1167565793 |
- |
- |
magnesium/proton exchanger, putative [Ricinus communis] |
| 10 |
Hb_000327_360 |
0.1240453285 |
- |
- |
unnamed protein product [Coffea canephora] |
| 11 |
Hb_000077_310 |
0.1284078755 |
- |
- |
PREDICTED: clathrin light chain 1 [Jatropha curcas] |
| 12 |
Hb_042083_030 |
0.1301004222 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 13 |
Hb_001180_010 |
0.1309859681 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 9 [Jatropha curcas] |
| 14 |
Hb_004041_050 |
0.1314458705 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 15 |
Hb_002397_050 |
0.1315827537 |
- |
- |
PREDICTED: ACT domain-containing protein ACR4 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000054_030 |
0.1331368374 |
- |
- |
hypothetical protein POPTR_0010s17680g [Populus trichocarpa] |
| 17 |
Hb_011174_040 |
0.1378611857 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_006736_050 |
0.138918808 |
- |
- |
hypothetical protein POPTR_0006s04450g [Populus trichocarpa] |
| 19 |
Hb_004032_110 |
0.1393514791 |
- |
- |
PREDICTED: putative methylesterase 14, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000115_360 |
0.1393947086 |
- |
- |
PREDICTED: probable protein phosphatase 2C 15 isoform X3 [Jatropha curcas] |