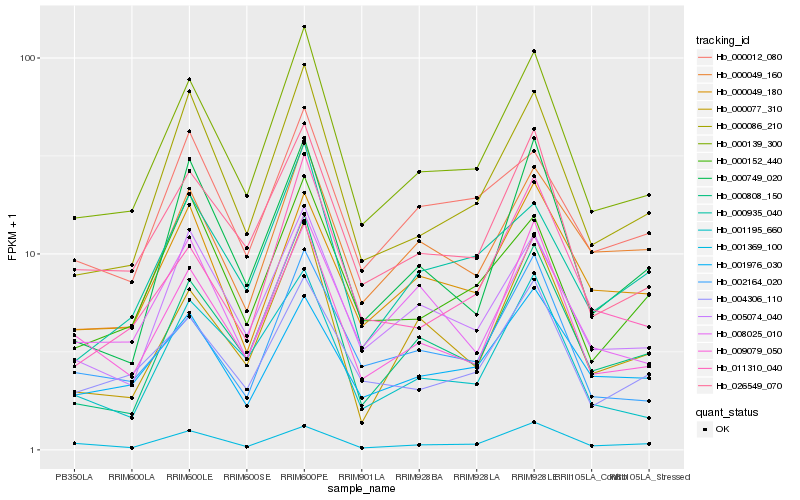
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000139_300 |
0.0 |
- |
- |
hypothetical protein POPTR_0008s19950g [Populus trichocarpa] |
| 2 |
Hb_000086_210 |
0.0710507435 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 3 |
Hb_026549_070 |
0.0714712211 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 1 [Jatropha curcas] |
| 4 |
Hb_005074_040 |
0.0818958678 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 5 |
Hb_004306_110 |
0.0903218282 |
transcription factor |
TF Family: Trihelix |
PREDICTED: phosphatidylinositol/phosphatidylcholine transfer protein SFH9 [Jatropha curcas] |
| 6 |
Hb_009079_050 |
0.0904889778 |
- |
- |
PREDICTED: V-type proton ATPase subunit a3-like [Jatropha curcas] |
| 7 |
Hb_000808_150 |
0.0951121143 |
- |
- |
PREDICTED: uncharacterized protein LOC105647444 [Jatropha curcas] |
| 8 |
Hb_011310_040 |
0.1024084093 |
transcription factor |
TF Family: bZIP |
transcription factor hy5, putative [Ricinus communis] |
| 9 |
Hb_001195_660 |
0.1059710614 |
- |
- |
metalloendopeptidase, putative [Ricinus communis] |
| 10 |
Hb_000935_040 |
0.106158043 |
- |
- |
Uncharacterized protein isoform 2 [Theobroma cacao] |
| 11 |
Hb_000152_440 |
0.1064434122 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 3-like [Jatropha curcas] |
| 12 |
Hb_002164_020 |
0.1075208554 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001976_030 |
0.1100604118 |
- |
- |
PREDICTED: protein NETWORKED 2D-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_000077_310 |
0.1114593421 |
- |
- |
PREDICTED: clathrin light chain 1 [Jatropha curcas] |
| 15 |
Hb_000049_160 |
0.1119076931 |
- |
- |
tyrosine decarboxylase family protein [Populus trichocarpa] |
| 16 |
Hb_000749_020 |
0.1131908777 |
- |
- |
Nucleotide-diphospho-sugar transferases superfamily protein isoform 1 [Theobroma cacao] |
| 17 |
Hb_008025_010 |
0.1144552429 |
- |
- |
PREDICTED: lysosomal beta glucosidase-like [Musa acuminata subsp. malaccensis] |
| 18 |
Hb_001369_100 |
0.1148340531 |
- |
- |
hypothetical protein JCGZ_26280 [Jatropha curcas] |
| 19 |
Hb_000049_180 |
0.1162286695 |
- |
- |
aromatic amino acid decarboxylase, putative [Ricinus communis] |
| 20 |
Hb_000012_080 |
0.1165720854 |
- |
- |
PREDICTED: clathrin interactor EPSIN 1 [Jatropha curcas] |