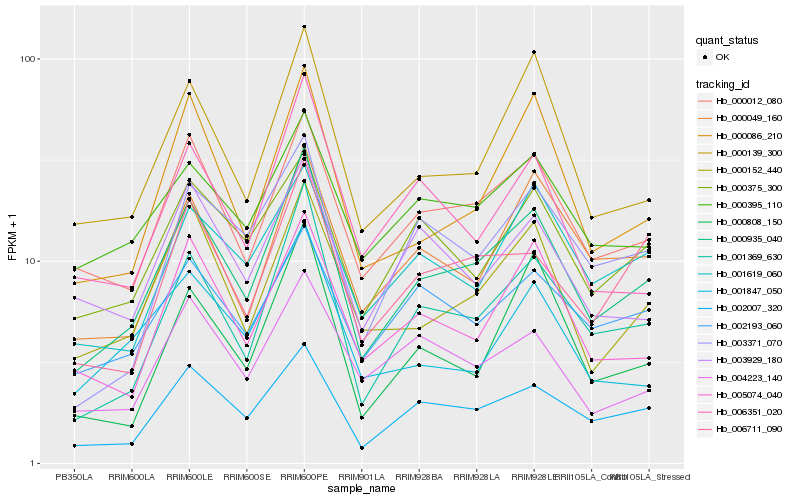
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000935_040 |
0.0 |
- |
- |
Uncharacterized protein isoform 2 [Theobroma cacao] |
| 2 |
Hb_001369_630 |
0.0807664476 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 3-like [Jatropha curcas] |
| 3 |
Hb_002007_320 |
0.09323892 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase ANP1 [Jatropha curcas] |
| 4 |
Hb_000012_080 |
0.0947266602 |
- |
- |
PREDICTED: clathrin interactor EPSIN 1 [Jatropha curcas] |
| 5 |
Hb_000152_440 |
0.1056135265 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 3-like [Jatropha curcas] |
| 6 |
Hb_000139_300 |
0.106158043 |
- |
- |
hypothetical protein POPTR_0008s19950g [Populus trichocarpa] |
| 7 |
Hb_000086_210 |
0.1102891176 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 8 |
Hb_005074_040 |
0.1165486993 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 9 |
Hb_000049_160 |
0.1204166246 |
- |
- |
tyrosine decarboxylase family protein [Populus trichocarpa] |
| 10 |
Hb_006711_090 |
0.1236965829 |
- |
- |
PREDICTED: fatty acid 2-hydroxylase 1-like [Jatropha curcas] |
| 11 |
Hb_000395_110 |
0.1255189653 |
- |
- |
PREDICTED: molybdate-anion transporter [Jatropha curcas] |
| 12 |
Hb_003371_070 |
0.1260155198 |
- |
- |
PREDICTED: citrate synthase, mitochondrial [Jatropha curcas] |
| 13 |
Hb_000375_300 |
0.1276015918 |
- |
- |
PREDICTED: cyanate hydratase [Jatropha curcas] |
| 14 |
Hb_001619_060 |
0.1285839607 |
- |
- |
PREDICTED: uncharacterized methyltransferase At1g78140, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000808_150 |
0.1290423311 |
- |
- |
PREDICTED: uncharacterized protein LOC105647444 [Jatropha curcas] |
| 16 |
Hb_004223_140 |
0.1309643018 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 17 |
Hb_002193_060 |
0.1320017663 |
- |
- |
PREDICTED: uncharacterized protein C4orf29 homolog [Pyrus x bretschneideri] |
| 18 |
Hb_001847_050 |
0.134209032 |
- |
- |
hypothetical protein CISIN_1g019843mg [Citrus sinensis] |
| 19 |
Hb_006351_020 |
0.134649911 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase S.4-like [Jatropha curcas] |
| 20 |
Hb_003929_180 |
0.1379195039 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g17430 isoform X1 [Glycine max] |