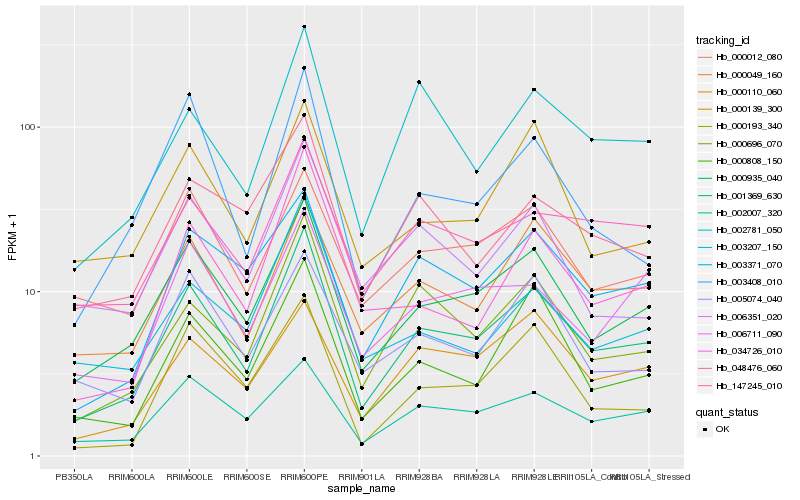
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001369_630 |
0.0 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 3-like [Jatropha curcas] |
| 2 |
Hb_000935_040 |
0.0807664476 |
- |
- |
Uncharacterized protein isoform 2 [Theobroma cacao] |
| 3 |
Hb_000696_070 |
0.1097125115 |
- |
- |
PREDICTED: aspartic proteinase-like protein 1 [Jatropha curcas] |
| 4 |
Hb_002007_320 |
0.1167229623 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase ANP1 [Jatropha curcas] |
| 5 |
Hb_000808_150 |
0.1177929185 |
- |
- |
PREDICTED: uncharacterized protein LOC105647444 [Jatropha curcas] |
| 6 |
Hb_003371_070 |
0.1214976918 |
- |
- |
PREDICTED: citrate synthase, mitochondrial [Jatropha curcas] |
| 7 |
Hb_000049_160 |
0.1221966622 |
- |
- |
tyrosine decarboxylase family protein [Populus trichocarpa] |
| 8 |
Hb_002781_050 |
0.127138795 |
- |
- |
cytochrome B5 isoform 1, putative [Ricinus communis] |
| 9 |
Hb_048476_060 |
0.1308081714 |
- |
- |
PREDICTED: CRAL-TRIO domain-containing protein YKL091C-like isoform X2 [Jatropha curcas] |
| 10 |
Hb_034726_010 |
0.1318118059 |
- |
- |
PREDICTED: chaperone protein dnaJ 1, mitochondrial isoform X2 [Jatropha curcas] |
| 11 |
Hb_000193_340 |
0.1350581411 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 isoform X1 [Jatropha curcas] |
| 12 |
Hb_147245_010 |
0.1352761956 |
- |
- |
casein kinase, putative [Ricinus communis] |
| 13 |
Hb_003207_150 |
0.1397942479 |
- |
- |
PREDICTED: SPX domain-containing membrane protein At4g22990-like [Jatropha curcas] |
| 14 |
Hb_000139_300 |
0.1411290914 |
- |
- |
hypothetical protein POPTR_0008s19950g [Populus trichocarpa] |
| 15 |
Hb_000110_060 |
0.1417326962 |
- |
- |
PREDICTED: uncharacterized protein LOC105636258 [Jatropha curcas] |
| 16 |
Hb_000012_080 |
0.1420428763 |
- |
- |
PREDICTED: clathrin interactor EPSIN 1 [Jatropha curcas] |
| 17 |
Hb_003408_010 |
0.1420503622 |
- |
- |
fructokinase [Manihot esculenta] |
| 18 |
Hb_006351_020 |
0.1431856274 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase S.4-like [Jatropha curcas] |
| 19 |
Hb_006711_090 |
0.1442161209 |
- |
- |
PREDICTED: fatty acid 2-hydroxylase 1-like [Jatropha curcas] |
| 20 |
Hb_005074_040 |
0.1450316081 |
- |
- |
ATP binding protein, putative [Ricinus communis] |