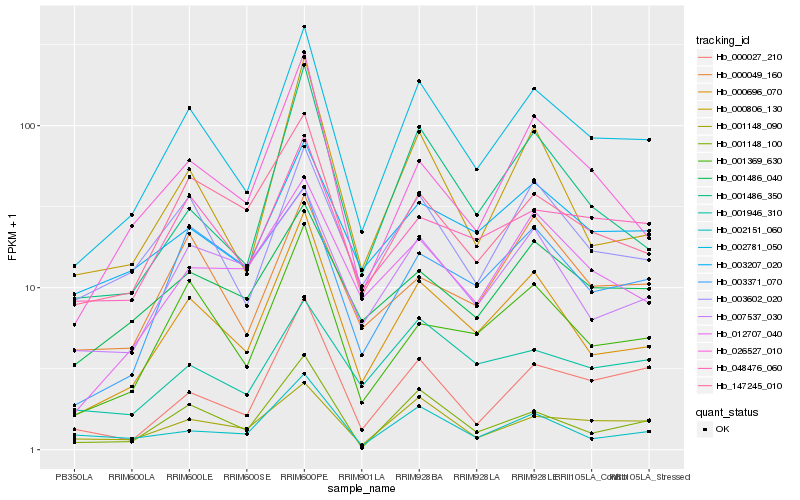
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002781_050 |
0.0 |
- |
- |
cytochrome B5 isoform 1, putative [Ricinus communis] |
| 2 |
Hb_000696_070 |
0.0899010146 |
- |
- |
PREDICTED: aspartic proteinase-like protein 1 [Jatropha curcas] |
| 3 |
Hb_003602_020 |
0.1171331888 |
- |
- |
PREDICTED: serine hydroxymethyltransferase, mitochondrial-like [Jatropha curcas] |
| 4 |
Hb_001148_100 |
0.1179331806 |
- |
- |
- |
| 5 |
Hb_001148_090 |
0.1235278153 |
- |
- |
hypothetical protein JCGZ_14711 [Jatropha curcas] |
| 6 |
Hb_001369_630 |
0.127138795 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 3-like [Jatropha curcas] |
| 7 |
Hb_048476_060 |
0.1307805686 |
- |
- |
PREDICTED: CRAL-TRIO domain-containing protein YKL091C-like isoform X2 [Jatropha curcas] |
| 8 |
Hb_000027_210 |
0.1317709272 |
- |
- |
Os01g0689300, partial [Oryza sativa Japonica Group] |
| 9 |
Hb_001486_350 |
0.1345122368 |
- |
- |
5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase, putative [Ricinus communis] |
| 10 |
Hb_003207_020 |
0.1347225456 |
- |
- |
PREDICTED: bifunctional dTDP-4-dehydrorhamnose 3,5-epimerase/dTDP-4-dehydrorhamnose reductase [Jatropha curcas] |
| 11 |
Hb_147245_010 |
0.1363862556 |
- |
- |
casein kinase, putative [Ricinus communis] |
| 12 |
Hb_000806_130 |
0.1393499545 |
- |
- |
Adenosine kinase 2 [Theobroma cacao] |
| 13 |
Hb_001486_040 |
0.1406314515 |
- |
- |
PREDICTED: uncharacterized protein LOC105632624 isoform X1 [Jatropha curcas] |
| 14 |
Hb_007537_030 |
0.1428374413 |
- |
- |
PREDICTED: gamma aminobutyrate transaminase 3, chloroplastic [Jatropha curcas] |
| 15 |
Hb_026527_010 |
0.1437133081 |
- |
- |
PREDICTED: caffeoylshikimate esterase-like [Populus euphratica] |
| 16 |
Hb_012707_040 |
0.1450907276 |
- |
- |
PREDICTED: probable inactive purple acid phosphatase 29 [Jatropha curcas] |
| 17 |
Hb_003371_070 |
0.1467648344 |
- |
- |
PREDICTED: citrate synthase, mitochondrial [Jatropha curcas] |
| 18 |
Hb_001946_310 |
0.1483245205 |
- |
- |
PREDICTED: probable ethanolamine kinase isoform X2 [Jatropha curcas] |
| 19 |
Hb_002151_060 |
0.1485546305 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit rpc4 [Jatropha curcas] |
| 20 |
Hb_000049_160 |
0.1500863499 |
- |
- |
tyrosine decarboxylase family protein [Populus trichocarpa] |