| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
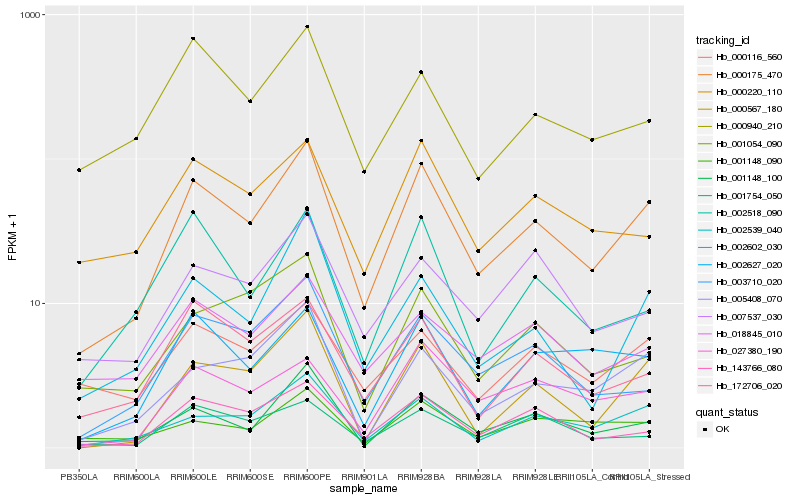
Hb_000175_470 |
0.0 |
- |
- |
lactoylglutathione lyase, putative [Ricinus communis] |
| 2 |
Hb_002539_040 |
0.1157339201 |
- |
- |
SPFH domain-containing protein 2 precursor, putative [Ricinus communis] |
| 3 |
Hb_001148_100 |
0.135350415 |
- |
- |
- |
| 4 |
Hb_143766_080 |
0.1424201639 |
- |
- |
PREDICTED: uncharacterized protein LOC105637797 isoform X1 [Jatropha curcas] |
| 5 |
Hb_005408_070 |
0.14552814 |
transcription factor |
TF Family: OFP |
PREDICTED: transcription repressor OFP4 [Jatropha curcas] |
| 6 |
Hb_172706_020 |
0.1455354855 |
- |
- |
PREDICTED: protein ABIL2 [Jatropha curcas] |
| 7 |
Hb_000567_180 |
0.1483273405 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF1.5 [Jatropha curcas] |
| 8 |
Hb_002627_020 |
0.149148408 |
- |
- |
PREDICTED: TBC1 domain family member 2B [Jatropha curcas] |
| 9 |
Hb_003710_020 |
0.1503997186 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53440 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000116_560 |
0.1510367637 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 [Jatropha curcas] |
| 11 |
Hb_002602_030 |
0.1516546627 |
- |
- |
PREDICTED: chaperone protein dnaJ 11, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000220_110 |
0.1519263512 |
- |
- |
fructose-bisphosphate aldolase, putative [Ricinus communis] |
| 13 |
Hb_001148_090 |
0.1519974047 |
- |
- |
hypothetical protein JCGZ_14711 [Jatropha curcas] |
| 14 |
Hb_027380_190 |
0.1527664759 |
- |
- |
PREDICTED: uncharacterized protein LOC105634028 isoform X1 [Jatropha curcas] |
| 15 |
Hb_018845_010 |
0.1586308782 |
- |
- |
PREDICTED: uncharacterized protein LOC105642055 [Jatropha curcas] |
| 16 |
Hb_002518_090 |
0.1595605251 |
- |
- |
PREDICTED: tubulin beta-2 chain [Jatropha curcas] |
| 17 |
Hb_000940_210 |
0.1642736519 |
- |
- |
cyclophilin [Hevea brasiliensis] |
| 18 |
Hb_007537_030 |
0.1644416318 |
- |
- |
PREDICTED: gamma aminobutyrate transaminase 3, chloroplastic [Jatropha curcas] |
| 19 |
Hb_001754_050 |
0.1650051069 |
- |
- |
protein arginine n-methyltransferase, putative [Ricinus communis] |
| 20 |
Hb_001054_090 |
0.1658167669 |
- |
- |
Potassium channel SKOR, putative [Ricinus communis] |