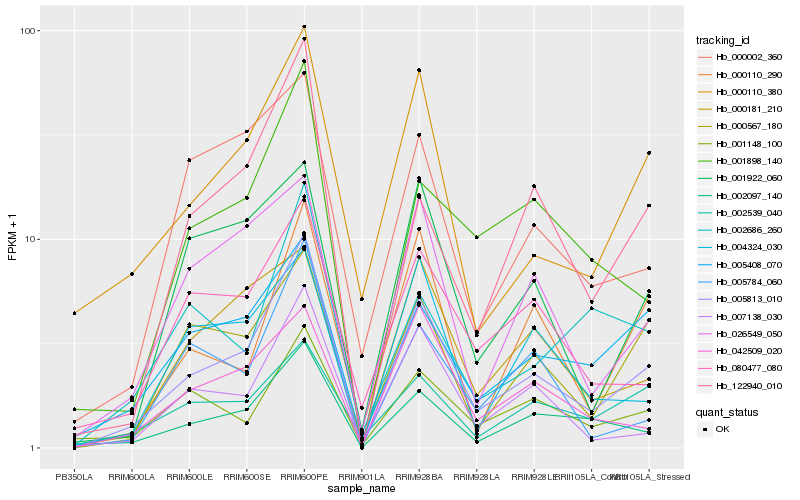
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005813_010 |
0.0 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 2 |
Hb_002097_140 |
0.1491256765 |
- |
- |
CHY zinc finger family protein, expressed [Oryza sativa Japonica Group] |
| 3 |
Hb_007138_030 |
0.1635845159 |
- |
- |
PREDICTED: putative disease resistance protein At4g11170 [Jatropha curcas] |
| 4 |
Hb_000110_380 |
0.1643268054 |
- |
- |
PREDICTED: uncharacterized protein LOC105642019 [Jatropha curcas] |
| 5 |
Hb_122940_010 |
0.1690892868 |
- |
- |
PREDICTED: 2-isopropylmalate synthase 2, chloroplastic-like [Populus euphratica] |
| 6 |
Hb_005408_070 |
0.17195952 |
transcription factor |
TF Family: OFP |
PREDICTED: transcription repressor OFP4 [Jatropha curcas] |
| 7 |
Hb_000567_180 |
0.1734276611 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF1.5 [Jatropha curcas] |
| 8 |
Hb_001898_140 |
0.1773127213 |
- |
- |
hypothetical protein AMTR_s00071p00184460 [Amborella trichopoda] |
| 9 |
Hb_001148_100 |
0.1798855769 |
- |
- |
- |
| 10 |
Hb_005784_060 |
0.1802988936 |
- |
- |
PREDICTED: U-box domain-containing protein 28 [Jatropha curcas] |
| 11 |
Hb_000181_210 |
0.1854969206 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 12 |
Hb_080477_080 |
0.1857843145 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4 isoform X1 [Gossypium raimondii] |
| 13 |
Hb_026549_050 |
0.1860791585 |
- |
- |
PREDICTED: uncharacterized protein LOC105636246 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000110_290 |
0.1877408369 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_042509_020 |
0.1892657861 |
- |
- |
hypothetical protein JCGZ_19555 [Jatropha curcas] |
| 16 |
Hb_001922_060 |
0.1932607146 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 17 |
Hb_004324_030 |
0.1967655907 |
- |
- |
PREDICTED: probable protein phosphatase 2C 8 [Jatropha curcas] |
| 18 |
Hb_002539_040 |
0.1967798829 |
- |
- |
SPFH domain-containing protein 2 precursor, putative [Ricinus communis] |
| 19 |
Hb_000002_360 |
0.1971653135 |
- |
- |
hypothetical protein POPTR_0008s16470g [Populus trichocarpa] |
| 20 |
Hb_002686_260 |
0.1997970493 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein 21/22, putative [Ricinus communis] |