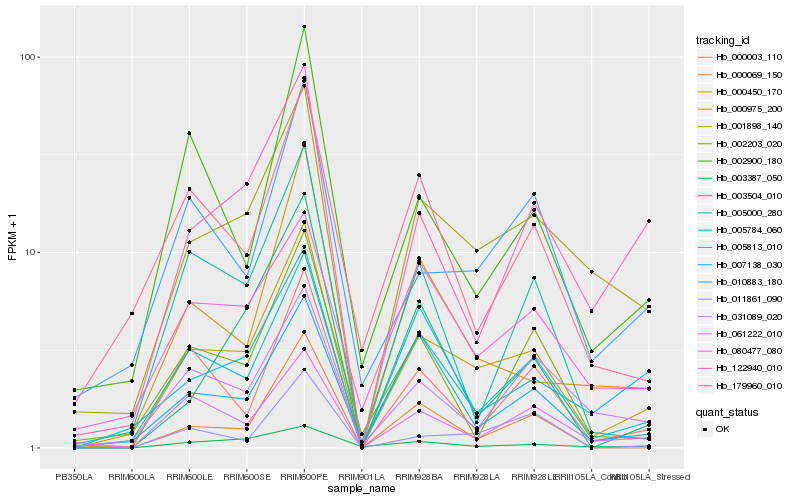
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000975_200 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105628231 isoform X2 [Jatropha curcas] |
| 2 |
Hb_005784_060 |
0.1212842716 |
- |
- |
PREDICTED: U-box domain-containing protein 28 [Jatropha curcas] |
| 3 |
Hb_001898_140 |
0.1367935356 |
- |
- |
hypothetical protein AMTR_s00071p00184460 [Amborella trichopoda] |
| 4 |
Hb_061222_010 |
0.1755873573 |
- |
- |
hypothetical protein RCOM_0012650 [Ricinus communis] |
| 5 |
Hb_000003_110 |
0.1757062437 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 6 |
Hb_000069_150 |
0.1759705328 |
- |
- |
PREDICTED: uncharacterized protein At4g00950 [Jatropha curcas] |
| 7 |
Hb_010883_180 |
0.1791258111 |
transcription factor |
TF Family: CSD |
PREDICTED: glycine-rich protein 2-like [Jatropha curcas] |
| 8 |
Hb_122940_010 |
0.1802655205 |
- |
- |
PREDICTED: 2-isopropylmalate synthase 2, chloroplastic-like [Populus euphratica] |
| 9 |
Hb_003387_050 |
0.1865885389 |
- |
- |
PREDICTED: uncharacterized protein LOC105639324 [Jatropha curcas] |
| 10 |
Hb_000450_170 |
0.1868184481 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_005000_280 |
0.1902293045 |
- |
- |
PREDICTED: uncharacterized protein LOC105637905 [Jatropha curcas] |
| 12 |
Hb_011861_090 |
0.1903752379 |
- |
- |
PREDICTED: probable sodium-coupled neutral amino acid transporter 6 [Jatropha curcas] |
| 13 |
Hb_031089_020 |
0.1914459149 |
- |
- |
Nodulation receptor kinase precursor, putative [Ricinus communis] |
| 14 |
Hb_002900_180 |
0.1938653857 |
transcription factor |
TF Family: LIM |
LIM1 [Hevea brasiliensis] |
| 15 |
Hb_007138_030 |
0.1953644577 |
- |
- |
PREDICTED: putative disease resistance protein At4g11170 [Jatropha curcas] |
| 16 |
Hb_002203_020 |
0.1996449086 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_080477_080 |
0.203017081 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4 isoform X1 [Gossypium raimondii] |
| 18 |
Hb_003504_010 |
0.2035343965 |
- |
- |
hypothetical protein POPTR_0013s04290g [Populus trichocarpa] |
| 19 |
Hb_179960_010 |
0.2048774024 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase ATL4-like [Jatropha curcas] |
| 20 |
Hb_005813_010 |
0.2054471935 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |