| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
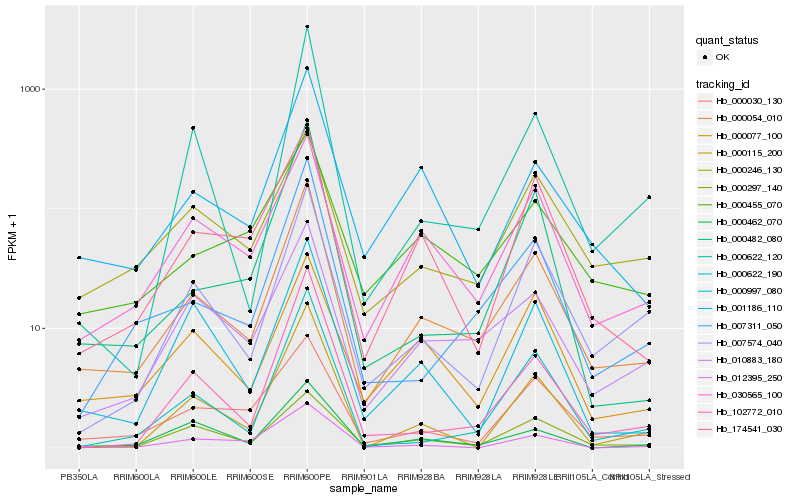
Hb_000054_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 2 |
Hb_030565_100 |
0.1287942723 |
- |
- |
serine hydroxymethyltransferase, putative [Ricinus communis] |
| 3 |
Hb_010883_180 |
0.1330153631 |
transcription factor |
TF Family: CSD |
PREDICTED: glycine-rich protein 2-like [Jatropha curcas] |
| 4 |
Hb_001186_110 |
0.1359715614 |
- |
- |
hypothetical protein JCGZ_18389 [Jatropha curcas] |
| 5 |
Hb_007311_050 |
0.1400080444 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_174541_030 |
0.1423850984 |
- |
- |
Cytochrome P450, family 98, subfamily A, polypeptide 3 [Theobroma cacao] |
| 7 |
Hb_007574_040 |
0.1428871306 |
- |
- |
PREDICTED: uncharacterized protein LOC105638144 [Jatropha curcas] |
| 8 |
Hb_000030_130 |
0.143713813 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 9 |
Hb_000297_140 |
0.1532346696 |
- |
- |
hypothetical protein RCOM_0293450 [Ricinus communis] |
| 10 |
Hb_000077_100 |
0.1588046427 |
- |
- |
PREDICTED: mannan endo-1,4-beta-mannosidase 2-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_000455_070 |
0.1595790071 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: carboxymethylenebutenolidase homolog [Jatropha curcas] |
| 12 |
Hb_000462_070 |
0.1604148126 |
- |
- |
PREDICTED: probable pectinesterase 15 [Jatropha curcas] |
| 13 |
Hb_000622_120 |
0.1605925617 |
- |
- |
hypothetical protein [Hevea brasiliensis] |
| 14 |
Hb_000622_190 |
0.16093436 |
- |
- |
PREDICTED: uncharacterized protein LOC105642890 [Jatropha curcas] |
| 15 |
Hb_000246_130 |
0.163028387 |
- |
- |
tubulin alpha chain, putative [Ricinus communis] |
| 16 |
Hb_000997_080 |
0.1670874781 |
- |
- |
PREDICTED: protein WVD2-like 1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_102772_010 |
0.1679099339 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000482_080 |
0.1692481944 |
- |
- |
PREDICTED: uncharacterized protein LOC105638144 [Jatropha curcas] |
| 19 |
Hb_000115_200 |
0.1696303963 |
- |
- |
PREDICTED: endonuclease 2 [Jatropha curcas] |
| 20 |
Hb_012395_250 |
0.1734048092 |
- |
- |
hypothetical protein RCOM_1217340 [Ricinus communis] |