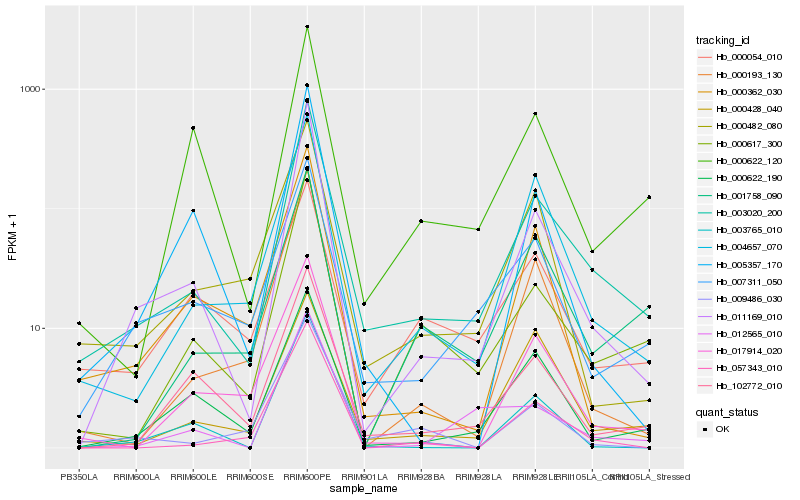
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003020_200 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105131117 [Populus euphratica] |
| 2 |
Hb_011169_010 |
0.1231337867 |
- |
- |
mta/sah nucleosidase, putative [Ricinus communis] |
| 3 |
Hb_000617_300 |
0.1423916026 |
- |
- |
RALFL33, putative [Ricinus communis] |
| 4 |
Hb_004657_070 |
0.1490312036 |
- |
- |
PREDICTED: protein IRX15-LIKE [Jatropha curcas] |
| 5 |
Hb_007311_050 |
0.1598270184 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_102772_010 |
0.1609249141 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000622_190 |
0.1657239358 |
- |
- |
PREDICTED: uncharacterized protein LOC105642890 [Jatropha curcas] |
| 8 |
Hb_000193_130 |
0.1663342973 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 9 |
Hb_000482_080 |
0.1709540947 |
- |
- |
PREDICTED: uncharacterized protein LOC105638144 [Jatropha curcas] |
| 10 |
Hb_017914_020 |
0.1736117571 |
- |
- |
- |
| 11 |
Hb_057343_010 |
0.1763093608 |
- |
- |
hypothetical protein JCGZ_23164 [Jatropha curcas] |
| 12 |
Hb_001758_090 |
0.1780733633 |
- |
- |
hypothetical protein POPTR_0006s09110g [Populus trichocarpa] |
| 13 |
Hb_000362_030 |
0.1797548372 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 14 |
Hb_003765_010 |
0.1824015997 |
- |
- |
berberine bridge enzyme [Hevea brasiliensis] |
| 15 |
Hb_009486_030 |
0.1847300291 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 12 [Jatropha curcas] |
| 16 |
Hb_000622_120 |
0.1861801078 |
- |
- |
hypothetical protein [Hevea brasiliensis] |
| 17 |
Hb_012565_010 |
0.1908737345 |
- |
- |
PREDICTED: putative glutamine amidotransferase PB2B2.05 [Jatropha curcas] |
| 18 |
Hb_000428_040 |
0.1920468843 |
- |
- |
PREDICTED: protein trichome birefringence-like 31 [Populus euphratica] |
| 19 |
Hb_005357_170 |
0.1927564461 |
- |
- |
Pectinesterase precursor, putative [Ricinus communis] |
| 20 |
Hb_000054_010 |
0.1944922013 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |