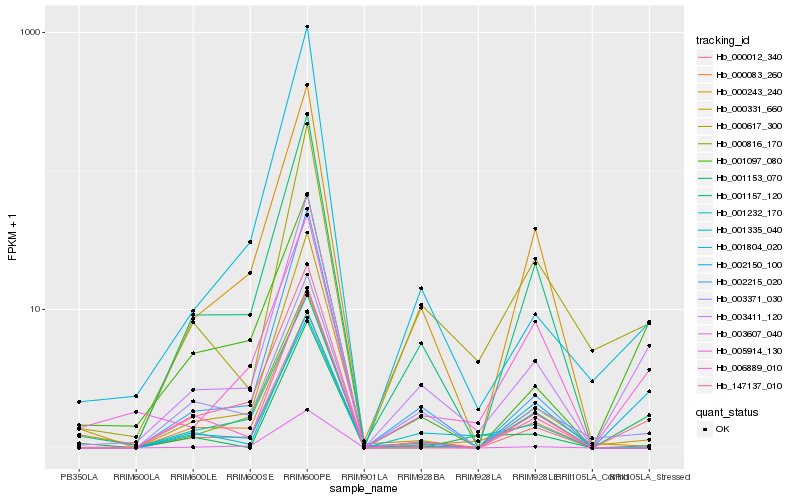
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003411_120 |
0.0 |
- |
- |
hypothetical protein POPTR_0010s13670g [Populus trichocarpa] |
| 2 |
Hb_001097_080 |
0.1472012454 |
transcription factor |
TF Family: MYB-related |
triptychon and cpc, putative [Ricinus communis] |
| 3 |
Hb_000617_300 |
0.1520962044 |
- |
- |
RALFL33, putative [Ricinus communis] |
| 4 |
Hb_147137_010 |
0.1545634915 |
- |
- |
hypothetical protein JCGZ_07229 [Jatropha curcas] |
| 5 |
Hb_003371_030 |
0.1627783315 |
- |
- |
PREDICTED: RING-H2 finger protein ATL5 isoform X1 [Jatropha curcas] |
| 6 |
Hb_005914_130 |
0.1719944753 |
- |
- |
- |
| 7 |
Hb_001804_020 |
0.1854692715 |
- |
- |
hypothetical protein POPTR_0006s09110g [Populus trichocarpa] |
| 8 |
Hb_001157_120 |
0.1855811187 |
- |
- |
PREDICTED: dirigent protein 19-like [Jatropha curcas] |
| 9 |
Hb_002215_020 |
0.1869099685 |
- |
- |
hypothetical protein JCGZ_13728 [Jatropha curcas] |
| 10 |
Hb_001153_070 |
0.1886143751 |
- |
- |
- |
| 11 |
Hb_002150_100 |
0.1886382688 |
- |
- |
PREDICTED: putative glucose-6-phosphate 1-epimerase isoform X1 [Jatropha curcas] |
| 12 |
Hb_001232_170 |
0.1893544002 |
- |
- |
sulfate transporter, putative [Ricinus communis] |
| 13 |
Hb_000243_240 |
0.1897113865 |
- |
- |
cytochrome P450 family protein [Populus trichocarpa] |
| 14 |
Hb_000331_660 |
0.1916296121 |
- |
- |
PREDICTED: vignain [Jatropha curcas] |
| 15 |
Hb_000083_260 |
0.1918995099 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000012_340 |
0.1958463296 |
- |
- |
PREDICTED: inorganic pyrophosphatase 3-like [Jatropha curcas] |
| 17 |
Hb_000816_170 |
0.1963882097 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 15 [Jatropha curcas] |
| 18 |
Hb_001335_040 |
0.19644056 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_003607_040 |
0.1979510048 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 20 |
Hb_006889_010 |
0.1985499472 |
- |
- |
PREDICTED: agmatine coumaroyltransferase-2-like [Jatropha curcas] |