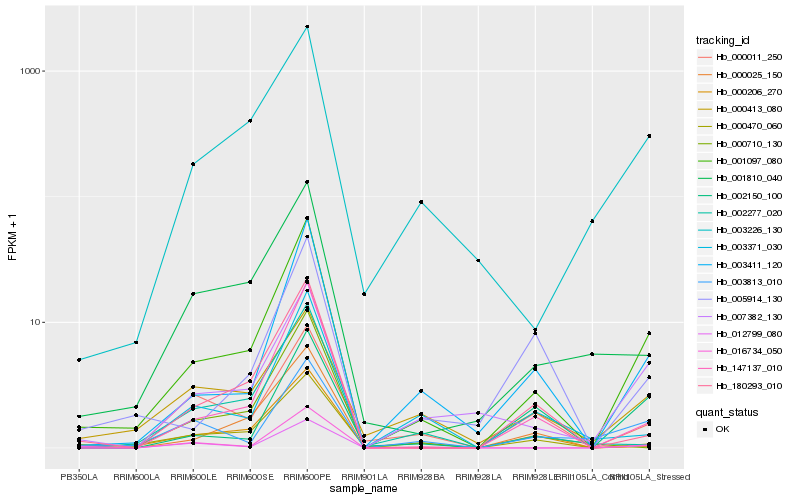
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001097_080 |
0.0 |
transcription factor |
TF Family: MYB-related |
triptychon and cpc, putative [Ricinus communis] |
| 2 |
Hb_003411_120 |
0.1472012454 |
- |
- |
hypothetical protein POPTR_0010s13670g [Populus trichocarpa] |
| 3 |
Hb_147137_010 |
0.1575199211 |
- |
- |
hypothetical protein JCGZ_07229 [Jatropha curcas] |
| 4 |
Hb_007382_130 |
0.1743401388 |
- |
- |
glycerophosphodiester phosphodiesterase, putative [Ricinus communis] |
| 5 |
Hb_000470_060 |
0.1853758284 |
- |
- |
PREDICTED: abietadienol/abietadienal oxidase [Jatropha curcas] |
| 6 |
Hb_012799_080 |
0.1894161625 |
transcription factor |
TF Family: MYB |
Myb domain protein 18, putative [Theobroma cacao] |
| 7 |
Hb_016734_050 |
0.1918048712 |
transcription factor |
TF Family: C2C2-GATA |
GATA transcription factor, putative [Ricinus communis] |
| 8 |
Hb_180293_010 |
0.1919079315 |
- |
- |
hypothetical protein POPTR_0008s19020g [Populus trichocarpa] |
| 9 |
Hb_003226_130 |
0.1972839498 |
- |
- |
PREDICTED: indole-3-acetic acid-induced protein ARG2-like [Jatropha curcas] |
| 10 |
Hb_003813_010 |
0.2042777402 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor BEE 1-like [Jatropha curcas] |
| 11 |
Hb_001810_040 |
0.204576705 |
- |
- |
- |
| 12 |
Hb_000206_270 |
0.2074897427 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 13 |
Hb_002277_020 |
0.210786806 |
- |
- |
Uncharacterized protein TCM_002409 [Theobroma cacao] |
| 14 |
Hb_003371_030 |
0.2111446475 |
- |
- |
PREDICTED: RING-H2 finger protein ATL5 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000710_130 |
0.2153666173 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 16 |
Hb_002150_100 |
0.2153696771 |
- |
- |
PREDICTED: putative glucose-6-phosphate 1-epimerase isoform X1 [Jatropha curcas] |
| 17 |
Hb_000413_080 |
0.2154523457 |
- |
- |
- |
| 18 |
Hb_000011_250 |
0.2183084494 |
- |
- |
PREDICTED: RING-H2 finger protein ATL3-like [Jatropha curcas] |
| 19 |
Hb_000025_150 |
0.2197286248 |
transcription factor |
TF Family: HSF |
hypothetical protein POPTR_0001s28040g [Populus trichocarpa] |
| 20 |
Hb_005914_130 |
0.2199910662 |
- |
- |
- |