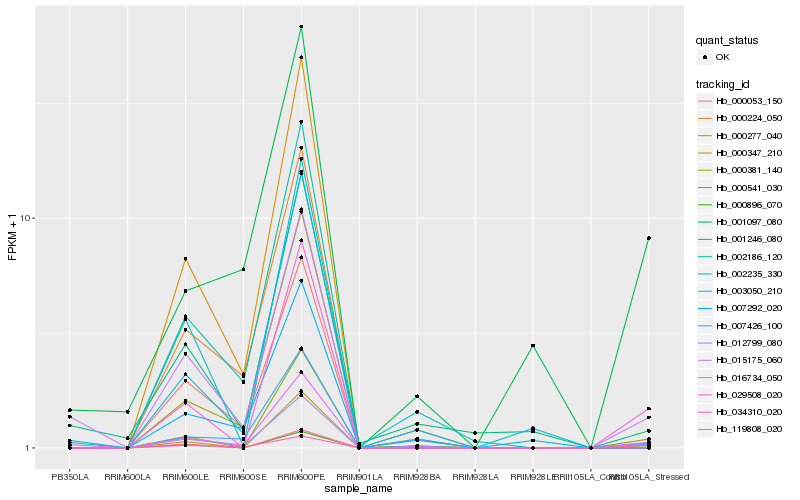
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012799_080 |
0.0 |
transcription factor |
TF Family: MYB |
Myb domain protein 18, putative [Theobroma cacao] |
| 2 |
Hb_000347_210 |
0.1220876111 |
- |
- |
PREDICTED: uncharacterized protein LOC105631371 [Jatropha curcas] |
| 3 |
Hb_016734_050 |
0.1573693264 |
transcription factor |
TF Family: C2C2-GATA |
GATA transcription factor, putative [Ricinus communis] |
| 4 |
Hb_029508_020 |
0.1614705934 |
- |
- |
- |
| 5 |
Hb_000541_030 |
0.1665145423 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_015175_060 |
0.1671668273 |
- |
- |
copper transport protein atox1, putative [Ricinus communis] |
| 7 |
Hb_119808_020 |
0.1710378235 |
- |
- |
PREDICTED: premnaspirodiene oxygenase-like [Vitis vinifera] |
| 8 |
Hb_000224_050 |
0.1743031421 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000896_070 |
0.1805846956 |
- |
- |
ammonium transporter, putative [Ricinus communis] |
| 10 |
Hb_001246_080 |
0.1826652977 |
- |
- |
PREDICTED: putative invertase inhibitor [Jatropha curcas] |
| 11 |
Hb_003050_210 |
0.1873962523 |
- |
- |
PREDICTED: cytochrome b561 domain-containing protein At4g18260-like [Populus euphratica] |
| 12 |
Hb_002235_330 |
0.1874170704 |
- |
- |
PREDICTED: expansin-A1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000381_140 |
0.1881678202 |
- |
- |
PREDICTED: S-type anion channel SLAH4-like [Jatropha curcas] |
| 14 |
Hb_001097_080 |
0.1894161625 |
transcription factor |
TF Family: MYB-related |
triptychon and cpc, putative [Ricinus communis] |
| 15 |
Hb_007292_020 |
0.1922434089 |
- |
- |
Indole-3-acetic acid-amido synthetase GH3.6, putative [Ricinus communis] |
| 16 |
Hb_000277_040 |
0.1927522141 |
- |
- |
- |
| 17 |
Hb_034310_020 |
0.1933020373 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK3 [Jatropha curcas] |
| 18 |
Hb_002186_120 |
0.1938433446 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53440 isoform X5 [Populus euphratica] |
| 19 |
Hb_000053_150 |
0.1940494203 |
- |
- |
clavata3/esr-related 12 family protein [Populus trichocarpa] |
| 20 |
Hb_007426_100 |
0.194136115 |
- |
- |
conserved hypothetical protein [Ricinus communis] |