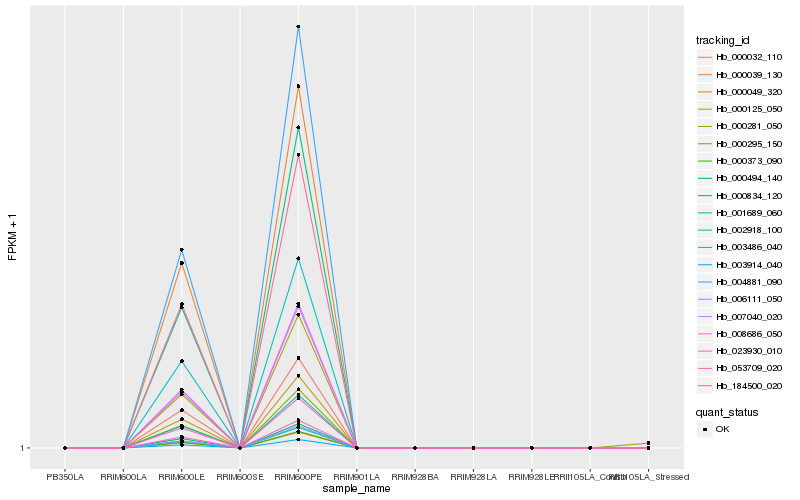
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000281_050 |
0.0 |
- |
- |
PREDICTED: subtilisin-like protease SBT3.5 isoform X2 [Populus euphratica] |
| 2 |
Hb_003914_040 |
0.1037247935 |
- |
- |
hypothetical protein EUGRSUZ_H02626 [Eucalyptus grandis] |
| 3 |
Hb_007040_020 |
0.1037250376 |
- |
- |
- |
| 4 |
Hb_000295_150 |
0.1037540171 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 5 |
Hb_000125_050 |
0.1040317477 |
- |
- |
- |
| 6 |
Hb_008686_050 |
0.1040319148 |
- |
- |
- |
| 7 |
Hb_000494_140 |
0.1041722947 |
transcription factor |
TF Family: ERF |
hypothetical protein JCGZ_14625 [Jatropha curcas] |
| 8 |
Hb_000039_130 |
0.1043997546 |
- |
- |
- |
| 9 |
Hb_000049_320 |
0.1045389647 |
- |
- |
PREDICTED: probable indole-3-pyruvate monooxygenase YUCCA5 [Jatropha curcas] |
| 10 |
Hb_053709_020 |
0.1046476652 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 11 |
Hb_184500_020 |
0.1046615221 |
- |
- |
- |
| 12 |
Hb_003486_040 |
0.1046640757 |
- |
- |
PREDICTED: glutamic acid-rich protein-like [Jatropha curcas] |
| 13 |
Hb_000373_090 |
0.1047111163 |
- |
- |
PREDICTED: abscisic acid 8'-hydroxylase 3-like [Prunus mume] |
| 14 |
Hb_023930_010 |
0.1048007287 |
- |
- |
hypothetical protein RCOM_0641040 [Ricinus communis] |
| 15 |
Hb_000834_120 |
0.1049137804 |
transcription factor |
TF Family: OFP |
PREDICTED: transcription repressor OFP8 [Populus euphratica] |
| 16 |
Hb_000032_110 |
0.1050858117 |
- |
- |
nadhp hc toxin reductase, putative [Ricinus communis] |
| 17 |
Hb_002918_100 |
0.1051052352 |
- |
- |
hypothetical protein JCGZ_21466 [Jatropha curcas] |
| 18 |
Hb_004881_090 |
0.1053207966 |
- |
- |
hypothetical protein POPTR_0002s06050g [Populus trichocarpa] |
| 19 |
Hb_001689_060 |
0.1058317182 |
- |
- |
PREDICTED: uncharacterized protein LOC103846154 [Brassica rapa] |
| 20 |
Hb_006111_050 |
0.1061526859 |
transcription factor |
TF Family: AP2 |
Protein AINTEGUMENTA, putative [Ricinus communis] |