| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
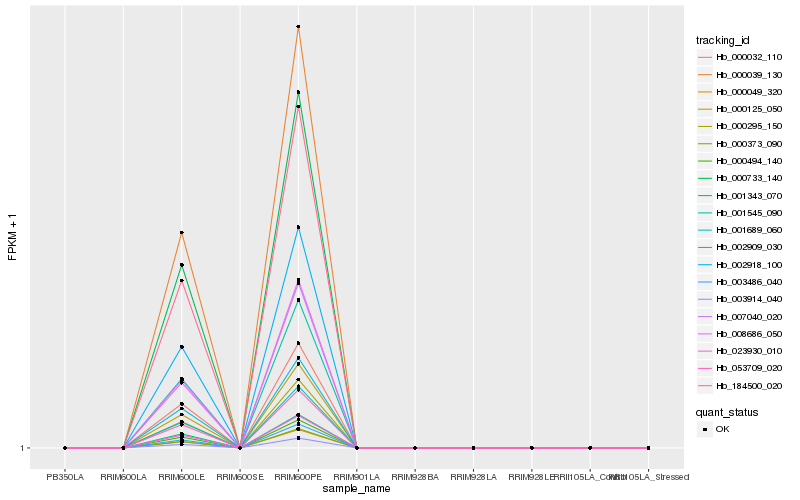
Hb_000494_140 |
0.0 |
transcription factor |
TF Family: ERF |
hypothetical protein JCGZ_14625 [Jatropha curcas] |
| 2 |
Hb_000125_050 |
0.0015583719 |
- |
- |
- |
| 3 |
Hb_000039_130 |
0.0021168195 |
- |
- |
- |
| 4 |
Hb_053709_020 |
0.0040767143 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 5 |
Hb_184500_020 |
0.0041785485 |
- |
- |
- |
| 6 |
Hb_023930_010 |
0.0051650578 |
- |
- |
hypothetical protein RCOM_0641040 [Ricinus communis] |
| 7 |
Hb_000295_150 |
0.0060790098 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 8 |
Hb_007040_020 |
0.0068805351 |
- |
- |
- |
| 9 |
Hb_003914_040 |
0.0068880924 |
- |
- |
hypothetical protein EUGRSUZ_H02626 [Eucalyptus grandis] |
| 10 |
Hb_000032_110 |
0.0070140706 |
- |
- |
nadhp hc toxin reductase, putative [Ricinus communis] |
| 11 |
Hb_002918_100 |
0.0071330766 |
- |
- |
hypothetical protein JCGZ_21466 [Jatropha curcas] |
| 12 |
Hb_001689_060 |
0.011126718 |
- |
- |
PREDICTED: uncharacterized protein LOC103846154 [Brassica rapa] |
| 13 |
Hb_000733_140 |
0.0134989329 |
- |
- |
PREDICTED: ras-related protein Rab2BV-like [Jatropha curcas] |
| 14 |
Hb_001343_070 |
0.017011687 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g62680, mitochondrial-like [Jatropha curcas] |
| 15 |
Hb_002909_030 |
0.0171936805 |
transcription factor |
TF Family: C2H2 |
hypothetical protein RCOM_1270600 [Ricinus communis] |
| 16 |
Hb_008686_050 |
0.0189850384 |
- |
- |
- |
| 17 |
Hb_001545_090 |
0.0218493206 |
- |
- |
PREDICTED: uncharacterized protein LOC103874823 [Brassica rapa] |
| 18 |
Hb_000049_320 |
0.0237914974 |
- |
- |
PREDICTED: probable indole-3-pyruvate monooxygenase YUCCA5 [Jatropha curcas] |
| 19 |
Hb_003486_040 |
0.0247365133 |
- |
- |
PREDICTED: glutamic acid-rich protein-like [Jatropha curcas] |
| 20 |
Hb_000373_090 |
0.0250765066 |
- |
- |
PREDICTED: abscisic acid 8'-hydroxylase 3-like [Prunus mume] |