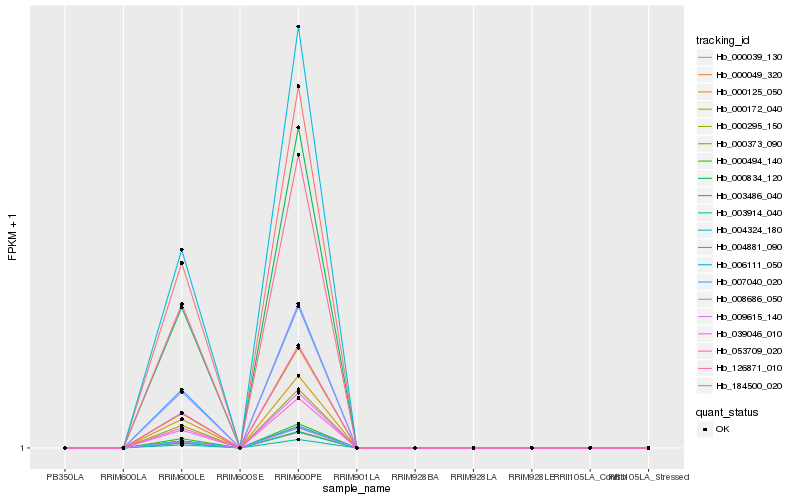
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008686_050 |
0.0 |
- |
- |
- |
| 2 |
Hb_000049_320 |
0.0048076733 |
- |
- |
PREDICTED: probable indole-3-pyruvate monooxygenase YUCCA5 [Jatropha curcas] |
| 3 |
Hb_003486_040 |
0.0057529878 |
- |
- |
PREDICTED: glutamic acid-rich protein-like [Jatropha curcas] |
| 4 |
Hb_000373_090 |
0.0060930933 |
- |
- |
PREDICTED: abscisic acid 8'-hydroxylase 3-like [Prunus mume] |
| 5 |
Hb_000834_120 |
0.0074784646 |
transcription factor |
TF Family: OFP |
PREDICTED: transcription repressor OFP8 [Populus euphratica] |
| 6 |
Hb_004881_090 |
0.0099594559 |
- |
- |
hypothetical protein POPTR_0002s06050g [Populus trichocarpa] |
| 7 |
Hb_003914_040 |
0.0120978101 |
- |
- |
hypothetical protein EUGRSUZ_H02626 [Eucalyptus grandis] |
| 8 |
Hb_007040_020 |
0.012105367 |
- |
- |
- |
| 9 |
Hb_000295_150 |
0.0129068414 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 10 |
Hb_006111_050 |
0.0142325626 |
transcription factor |
TF Family: AP2 |
Protein AINTEGUMENTA, putative [Ricinus communis] |
| 11 |
Hb_000172_040 |
0.0163131666 |
- |
- |
Tetratricopeptide repeat protein, putative [Ricinus communis] |
| 12 |
Hb_126871_010 |
0.016738829 |
- |
- |
PREDICTED: probable protein phosphatase 2C 38 [Jatropha curcas] |
| 13 |
Hb_009615_140 |
0.0170527322 |
- |
- |
PREDICTED: alpha-galactosidase-like isoform X2 [Pyrus x bretschneideri] |
| 14 |
Hb_000125_050 |
0.017426946 |
- |
- |
- |
| 15 |
Hb_039046_010 |
0.0186988824 |
- |
- |
PREDICTED: uncharacterized protein LOC105644040 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000494_140 |
0.0189850384 |
transcription factor |
TF Family: ERF |
hypothetical protein JCGZ_14625 [Jatropha curcas] |
| 17 |
Hb_000039_130 |
0.0211014004 |
- |
- |
- |
| 18 |
Hb_053709_020 |
0.0230607923 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 19 |
Hb_184500_020 |
0.0231625984 |
- |
- |
- |
| 20 |
Hb_004324_180 |
0.0239160652 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |