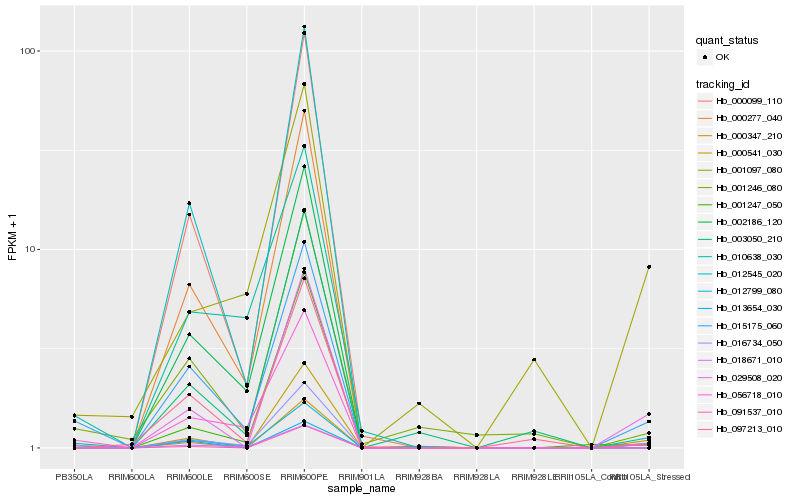
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_016734_050 |
0.0 |
transcription factor |
TF Family: C2C2-GATA |
GATA transcription factor, putative [Ricinus communis] |
| 2 |
Hb_015175_060 |
0.0781814146 |
- |
- |
copper transport protein atox1, putative [Ricinus communis] |
| 3 |
Hb_000541_030 |
0.1520244696 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_029508_020 |
0.152715523 |
- |
- |
- |
| 5 |
Hb_012799_080 |
0.1573693264 |
transcription factor |
TF Family: MYB |
Myb domain protein 18, putative [Theobroma cacao] |
| 6 |
Hb_056718_010 |
0.1691477725 |
- |
- |
hypothetical protein JCGZ_11777 [Jatropha curcas] |
| 7 |
Hb_001246_080 |
0.1802614424 |
- |
- |
PREDICTED: putative invertase inhibitor [Jatropha curcas] |
| 8 |
Hb_000347_210 |
0.1835996907 |
- |
- |
PREDICTED: uncharacterized protein LOC105631371 [Jatropha curcas] |
| 9 |
Hb_013654_030 |
0.186387342 |
- |
- |
Dehydration-responsive protein RD22 precursor, putative [Ricinus communis] |
| 10 |
Hb_000277_040 |
0.1878877601 |
- |
- |
- |
| 11 |
Hb_002186_120 |
0.1888580413 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53440 isoform X5 [Populus euphratica] |
| 12 |
Hb_003050_210 |
0.1896134976 |
- |
- |
PREDICTED: cytochrome b561 domain-containing protein At4g18260-like [Populus euphratica] |
| 13 |
Hb_018671_010 |
0.1898798061 |
- |
- |
- |
| 14 |
Hb_001247_050 |
0.190817983 |
- |
- |
PREDICTED: WAT1-related protein At1g09380 [Jatropha curcas] |
| 15 |
Hb_091537_010 |
0.1914421038 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 16 |
Hb_010638_030 |
0.1917613815 |
- |
- |
oleosin1 [Plukenetia volubilis] |
| 17 |
Hb_001097_080 |
0.1918048712 |
transcription factor |
TF Family: MYB-related |
triptychon and cpc, putative [Ricinus communis] |
| 18 |
Hb_000099_110 |
0.1951676647 |
- |
- |
PREDICTED: disease resistance response protein 206-like [Jatropha curcas] |
| 19 |
Hb_012545_020 |
0.1960725129 |
- |
- |
PREDICTED: uncharacterized protein LOC105633903 [Jatropha curcas] |
| 20 |
Hb_097213_010 |
0.1968035886 |
- |
- |
PREDICTED: cytochrome P450 714C2-like [Populus euphratica] |