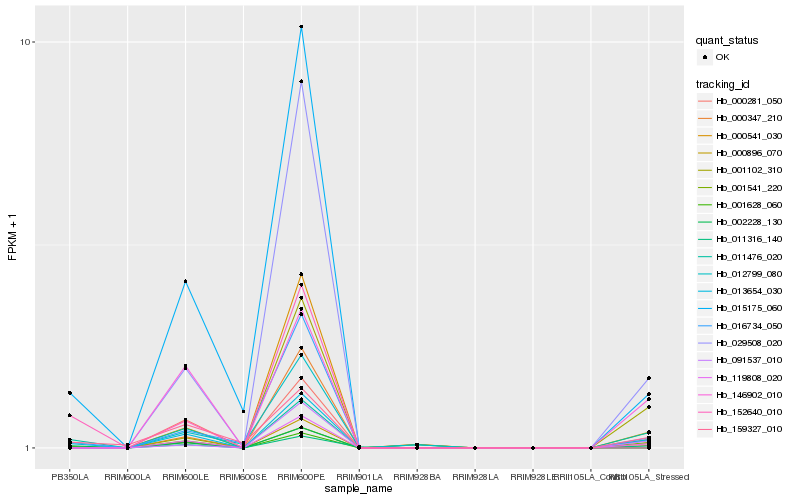
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_013654_030 |
0.0 |
- |
- |
Dehydration-responsive protein RD22 precursor, putative [Ricinus communis] |
| 2 |
Hb_119808_020 |
0.158548218 |
- |
- |
PREDICTED: premnaspirodiene oxygenase-like [Vitis vinifera] |
| 3 |
Hb_015175_060 |
0.1658686551 |
- |
- |
copper transport protein atox1, putative [Ricinus communis] |
| 4 |
Hb_011476_020 |
0.1827514052 |
- |
- |
PREDICTED: glutamate dehydrogenase 2-like [Jatropha curcas] |
| 5 |
Hb_001541_220 |
0.1860746149 |
transcription factor |
TF Family: ERF |
hypothetical protein RCOM_0793640 [Ricinus communis] |
| 6 |
Hb_016734_050 |
0.186387342 |
transcription factor |
TF Family: C2C2-GATA |
GATA transcription factor, putative [Ricinus communis] |
| 7 |
Hb_159327_010 |
0.1972259017 |
- |
- |
hypothetical protein JCGZ_23768 [Jatropha curcas] |
| 8 |
Hb_029508_020 |
0.2138883141 |
- |
- |
- |
| 9 |
Hb_146902_010 |
0.2160634821 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002228_130 |
0.2235871552 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000281_050 |
0.2248955198 |
- |
- |
PREDICTED: subtilisin-like protease SBT3.5 isoform X2 [Populus euphratica] |
| 12 |
Hb_091537_010 |
0.227746032 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 13 |
Hb_000541_030 |
0.2283257815 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_011316_140 |
0.2319403542 |
- |
- |
PREDICTED: uncharacterized protein LOC105129591 [Populus euphratica] |
| 15 |
Hb_000347_210 |
0.2366311596 |
- |
- |
PREDICTED: uncharacterized protein LOC105631371 [Jatropha curcas] |
| 16 |
Hb_012799_080 |
0.2379972561 |
transcription factor |
TF Family: MYB |
Myb domain protein 18, putative [Theobroma cacao] |
| 17 |
Hb_001628_060 |
0.2384956986 |
- |
- |
PREDICTED: beta-galactosidase 13-like isoform X2 [Jatropha curcas] |
| 18 |
Hb_001102_310 |
0.2515268533 |
- |
- |
Protein AFR, putative [Ricinus communis] |
| 19 |
Hb_000896_070 |
0.2537811213 |
- |
- |
ammonium transporter, putative [Ricinus communis] |
| 20 |
Hb_152640_010 |
0.2549703635 |
- |
- |
PREDICTED: disease resistance RPP13-like protein 4 [Populus euphratica] |