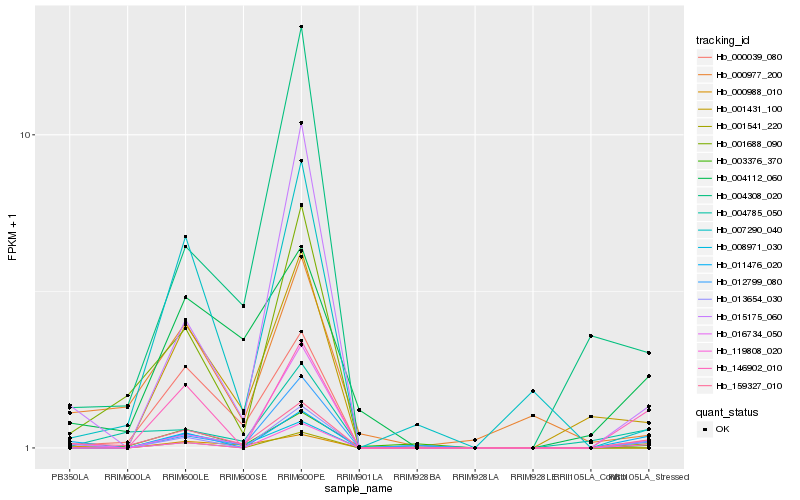
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_159327_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_23768 [Jatropha curcas] |
| 2 |
Hb_013654_030 |
0.1972259017 |
- |
- |
Dehydration-responsive protein RD22 precursor, putative [Ricinus communis] |
| 3 |
Hb_008971_030 |
0.2092692725 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_015175_060 |
0.2177089428 |
- |
- |
copper transport protein atox1, putative [Ricinus communis] |
| 5 |
Hb_004308_020 |
0.2383094866 |
- |
- |
hypothetical protein POPTR_0005s15590g [Populus trichocarpa] |
| 6 |
Hb_011476_020 |
0.2426206827 |
- |
- |
PREDICTED: glutamate dehydrogenase 2-like [Jatropha curcas] |
| 7 |
Hb_004785_050 |
0.2484541066 |
- |
- |
PREDICTED: serine decarboxylase [Jatropha curcas] |
| 8 |
Hb_016734_050 |
0.2490365836 |
transcription factor |
TF Family: C2C2-GATA |
GATA transcription factor, putative [Ricinus communis] |
| 9 |
Hb_001541_220 |
0.2553394806 |
transcription factor |
TF Family: ERF |
hypothetical protein RCOM_0793640 [Ricinus communis] |
| 10 |
Hb_004112_060 |
0.2553693451 |
- |
- |
hypothetical protein POPTR_0017s01950g, partial [Populus trichocarpa] |
| 11 |
Hb_001688_090 |
0.2573339315 |
- |
- |
alcohol dehydrogenase 7 [Vitis vinifera] |
| 12 |
Hb_146902_010 |
0.2590547296 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_119808_020 |
0.2601381968 |
- |
- |
PREDICTED: premnaspirodiene oxygenase-like [Vitis vinifera] |
| 14 |
Hb_000977_200 |
0.2639927928 |
- |
- |
PREDICTED: acyl-[acyl-carrier-protein] desaturase, chloroplastic-like [Jatropha curcas] |
| 15 |
Hb_001431_100 |
0.268017915 |
- |
- |
PREDICTED: probable protein phosphatase 2C 27 [Jatropha curcas] |
| 16 |
Hb_000988_010 |
0.270877359 |
- |
- |
hypothetical protein POPTR_0004s17730g [Populus trichocarpa] |
| 17 |
Hb_000039_080 |
0.2719345951 |
- |
- |
cyclin d, putative [Ricinus communis] |
| 18 |
Hb_003376_370 |
0.2719908322 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 19 |
Hb_012799_080 |
0.2742802861 |
transcription factor |
TF Family: MYB |
Myb domain protein 18, putative [Theobroma cacao] |
| 20 |
Hb_007290_040 |
0.274520351 |
- |
- |
PREDICTED: histidine-containing phosphotransfer protein 1-like [Jatropha curcas] |