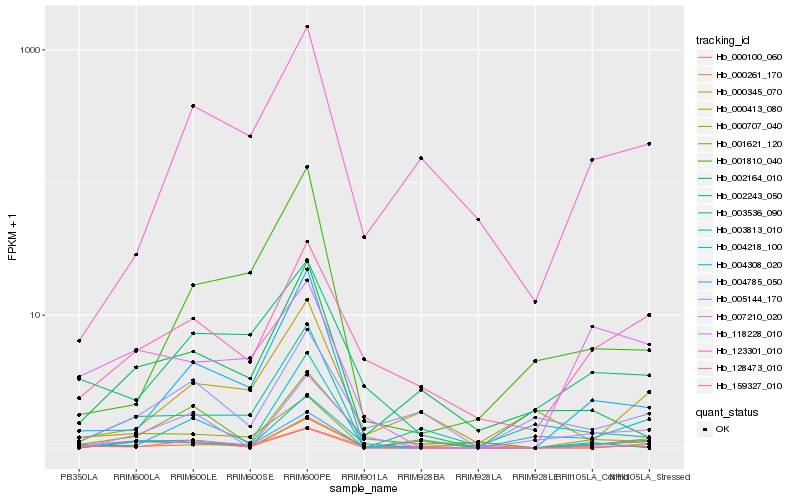
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004785_050 |
0.0 |
- |
- |
PREDICTED: serine decarboxylase [Jatropha curcas] |
| 2 |
Hb_004308_020 |
0.2156304276 |
- |
- |
hypothetical protein POPTR_0005s15590g [Populus trichocarpa] |
| 3 |
Hb_128473_010 |
0.2158104344 |
- |
- |
- |
| 4 |
Hb_000261_170 |
0.2411740055 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 5 |
Hb_159327_010 |
0.2484541066 |
- |
- |
hypothetical protein JCGZ_23768 [Jatropha curcas] |
| 6 |
Hb_007210_020 |
0.2533685818 |
- |
- |
PREDICTED: hexaprenyldihydroxybenzoate methyltransferase, mitochondrial-like [Solanum tuberosum] |
| 7 |
Hb_003813_010 |
0.2570225515 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor BEE 1-like [Jatropha curcas] |
| 8 |
Hb_000707_040 |
0.266005435 |
- |
- |
Uncharacterized protein TCM_021829 [Theobroma cacao] |
| 9 |
Hb_118228_010 |
0.2717355566 |
- |
- |
PREDICTED: pirin-like protein [Jatropha curcas] |
| 10 |
Hb_000100_060 |
0.2728411124 |
- |
- |
PREDICTED: probable glucan endo-1,3-beta-glucosidase A6 [Populus euphratica] |
| 11 |
Hb_002243_050 |
0.2765947201 |
- |
- |
PREDICTED: ion channel DMI1-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_001621_120 |
0.2774792664 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |
| 13 |
Hb_003536_090 |
0.2784762825 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 14 |
Hb_000345_070 |
0.2817561385 |
rubber biosynthesis |
Gene Name: SRPP6 |
- |
| 15 |
Hb_123301_010 |
0.2838748923 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_005144_170 |
0.2844232624 |
- |
- |
PREDICTED: shugoshin-1-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_004218_100 |
0.2844321418 |
- |
- |
PREDICTED: DNA-directed RNA polymerase V subunit 5C [Jatropha curcas] |
| 18 |
Hb_001810_040 |
0.2850214783 |
- |
- |
- |
| 19 |
Hb_000413_080 |
0.2866165795 |
- |
- |
- |
| 20 |
Hb_002164_010 |
0.2870796077 |
- |
- |
Stellacyanin, putative [Ricinus communis] |