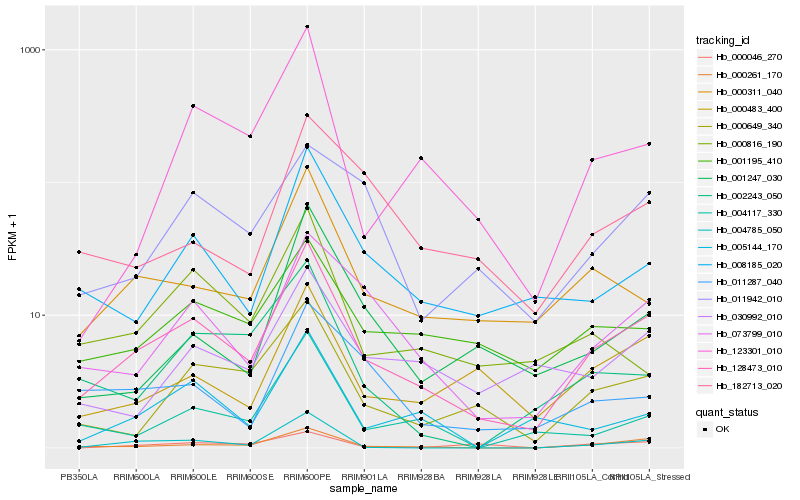
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_128473_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_000261_170 |
0.1612420496 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 3 |
Hb_073799_010 |
0.1758980905 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000649_340 |
0.1841094532 |
- |
- |
PREDICTED: dynein light chain 1, cytoplasmic [Jatropha curcas] |
| 5 |
Hb_000311_040 |
0.18958802 |
- |
- |
PREDICTED: S-formylglutathione hydrolase [Jatropha curcas] |
| 6 |
Hb_008185_020 |
0.1937400042 |
- |
- |
RING-H2 finger protein ATL3J, putative [Ricinus communis] |
| 7 |
Hb_000483_400 |
0.2006361045 |
- |
- |
PREDICTED: succinate dehydrogenase assembly factor 1, mitochondrial [Jatropha curcas] |
| 8 |
Hb_001195_410 |
0.2006781665 |
- |
- |
PREDICTED: deoxyuridine 5'-triphosphate nucleotidohydrolase [Jatropha curcas] |
| 9 |
Hb_123301_010 |
0.2102988039 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_004785_050 |
0.2158104344 |
- |
- |
PREDICTED: serine decarboxylase [Jatropha curcas] |
| 11 |
Hb_004117_330 |
0.2160801008 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_005144_170 |
0.2181592397 |
- |
- |
PREDICTED: shugoshin-1-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_030992_010 |
0.2185411538 |
- |
- |
acetylornithine aminotransferase, putative [Ricinus communis] |
| 14 |
Hb_002243_050 |
0.2189109181 |
- |
- |
PREDICTED: ion channel DMI1-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_000816_190 |
0.2202741936 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_011287_040 |
0.2240777297 |
- |
- |
PREDICTED: OTU domain-containing protein At3g57810-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_011942_010 |
0.2241503437 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001247_030 |
0.2255091248 |
- |
- |
- |
| 19 |
Hb_182713_020 |
0.2287542989 |
- |
- |
PREDICTED: vesicle-associated membrane protein 727 [Setaria italica] |
| 20 |
Hb_000046_270 |
0.2300828539 |
- |
- |
PREDICTED: sodium/hydrogen exchanger 2-like isoform X2 [Jatropha curcas] |