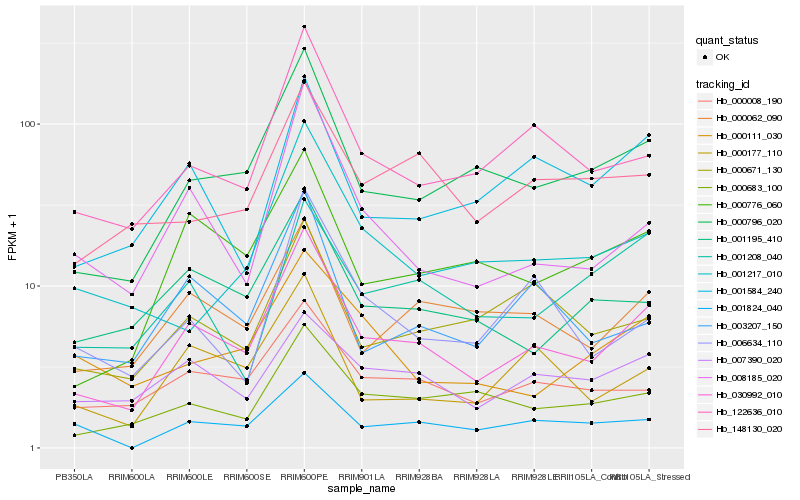
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_030992_010 |
0.0 |
- |
- |
acetylornithine aminotransferase, putative [Ricinus communis] |
| 2 |
Hb_000796_020 |
0.12494488 |
- |
- |
ADP-ribosylation factor [Phaseolus vulgaris] |
| 3 |
Hb_122636_010 |
0.1390255069 |
- |
- |
dienelactone hydrolase family protein [Populus trichocarpa] |
| 4 |
Hb_000776_060 |
0.1420676177 |
- |
- |
histone H4 [Zea mays] |
| 5 |
Hb_000062_090 |
0.1458018278 |
- |
- |
PREDICTED: uncharacterized protein LOC105644743 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000008_190 |
0.147359492 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000177_110 |
0.1525393999 |
- |
- |
PREDICTED: uncharacterized protein LOC105642063 [Jatropha curcas] |
| 8 |
Hb_000683_100 |
0.1545870777 |
- |
- |
hypothetical protein JCGZ_22137 [Jatropha curcas] |
| 9 |
Hb_003207_150 |
0.1568171471 |
- |
- |
PREDICTED: SPX domain-containing membrane protein At4g22990-like [Jatropha curcas] |
| 10 |
Hb_008185_020 |
0.1591405114 |
- |
- |
RING-H2 finger protein ATL3J, putative [Ricinus communis] |
| 11 |
Hb_001584_240 |
0.1600997357 |
- |
- |
ADP-ribosylation factor [Gossypium barbadense] |
| 12 |
Hb_001195_410 |
0.1611414351 |
- |
- |
PREDICTED: deoxyuridine 5'-triphosphate nucleotidohydrolase [Jatropha curcas] |
| 13 |
Hb_007390_020 |
0.165447271 |
- |
- |
PREDICTED: protein FMP32, mitochondrial-like [Populus euphratica] |
| 14 |
Hb_000671_130 |
0.1694356006 |
- |
- |
PREDICTED: CMP-sialic acid transporter 2 isoform X2 [Jatropha curcas] |
| 15 |
Hb_001824_040 |
0.1725025864 |
- |
- |
hypothetical protein M569_11359, partial [Genlisea aurea] |
| 16 |
Hb_148130_020 |
0.172948735 |
- |
- |
- |
| 17 |
Hb_006634_110 |
0.1770926007 |
- |
- |
BnaC04g40780D [Brassica napus] |
| 18 |
Hb_001217_010 |
0.1789904898 |
- |
- |
hypothetical protein JCGZ_04022 [Jatropha curcas] |
| 19 |
Hb_000111_030 |
0.1814028804 |
- |
- |
PREDICTED: uncharacterized protein LOC104094030 [Nicotiana tomentosiformis] |
| 20 |
Hb_001208_040 |
0.1817685859 |
- |
- |
hypothetical protein CISIN_1g025613mg [Citrus sinensis] |