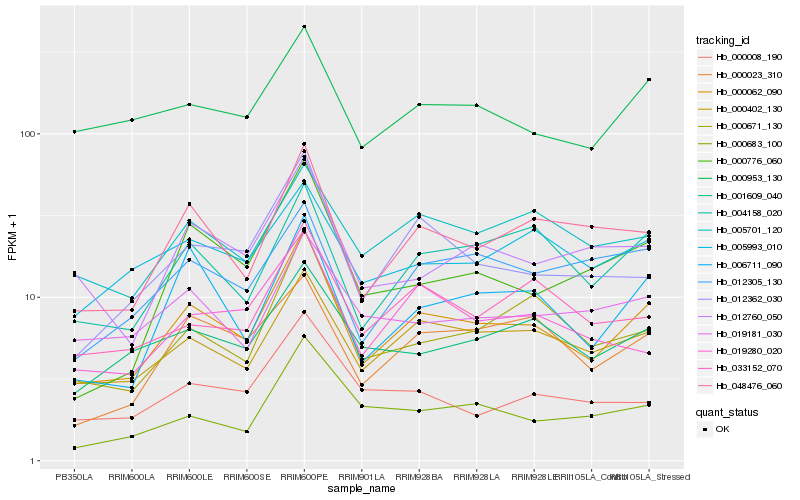
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000062_090 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644743 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000402_130 |
0.1057386336 |
- |
- |
PREDICTED: AP-1 complex subunit mu-2 [Jatropha curcas] |
| 3 |
Hb_048476_060 |
0.1129908107 |
- |
- |
PREDICTED: CRAL-TRIO domain-containing protein YKL091C-like isoform X2 [Jatropha curcas] |
| 4 |
Hb_006711_090 |
0.1170731423 |
- |
- |
PREDICTED: fatty acid 2-hydroxylase 1-like [Jatropha curcas] |
| 5 |
Hb_004158_020 |
0.1192241221 |
- |
- |
Rab1 [Hevea brasiliensis] |
| 6 |
Hb_000671_130 |
0.1223080386 |
- |
- |
PREDICTED: CMP-sialic acid transporter 2 isoform X2 [Jatropha curcas] |
| 7 |
Hb_012362_030 |
0.1227843959 |
- |
- |
PREDICTED: GPI-anchored protein LORELEI-like [Populus euphratica] |
| 8 |
Hb_000023_310 |
0.127557187 |
- |
- |
PREDICTED: probable folate-biopterin transporter 2 [Populus euphratica] |
| 9 |
Hb_001609_040 |
0.1315355436 |
- |
- |
hypothetical protein POPTR_0017s05650g [Populus trichocarpa] |
| 10 |
Hb_000953_130 |
0.1319057615 |
- |
- |
PREDICTED: fructose-bisphosphate aldolase cytoplasmic isozyme [Jatropha curcas] |
| 11 |
Hb_033152_070 |
0.1340497917 |
- |
- |
N-acetyl-gamma-glutamyl-phosphate reductase, putative [Ricinus communis] |
| 12 |
Hb_012760_050 |
0.13438305 |
- |
- |
RING-H2 finger protein ATL4M, putative [Ricinus communis] |
| 13 |
Hb_000776_060 |
0.1357744299 |
- |
- |
histone H4 [Zea mays] |
| 14 |
Hb_019280_020 |
0.1371512419 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK14 isoform X1 [Jatropha curcas] |
| 15 |
Hb_005701_120 |
0.1373279591 |
- |
- |
PREDICTED: V-type proton ATPase 16 kDa proteolipid subunit [Cucumis sativus] |
| 16 |
Hb_000683_100 |
0.1375197591 |
- |
- |
hypothetical protein JCGZ_22137 [Jatropha curcas] |
| 17 |
Hb_005993_010 |
0.1393378779 |
- |
- |
PREDICTED: protein transport protein SEC13 homolog B [Jatropha curcas] |
| 18 |
Hb_000008_190 |
0.1400935376 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 isoform X2 [Jatropha curcas] |
| 19 |
Hb_019181_030 |
0.1412861157 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: conserved oligomeric Golgi complex subunit 6-like [Pyrus x bretschneideri] |
| 20 |
Hb_012305_130 |
0.1416466306 |
- |
- |
PREDICTED: GTP-binding protein SAR1A [Jatropha curcas] |