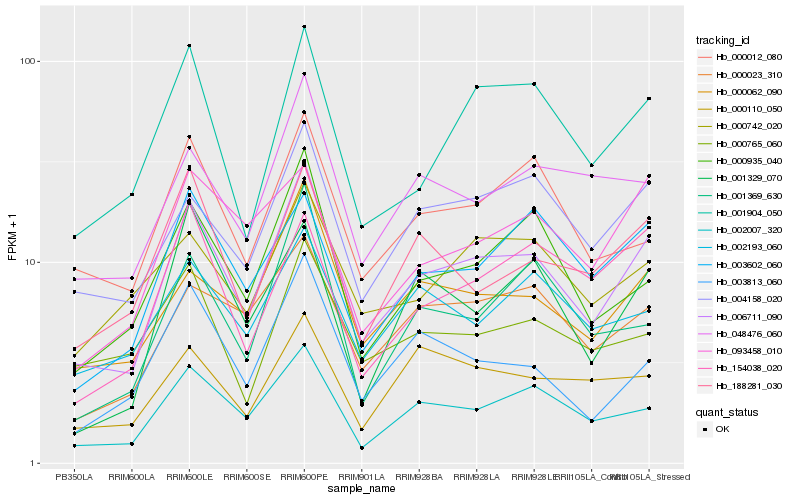
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006711_090 |
0.0 |
- |
- |
PREDICTED: fatty acid 2-hydroxylase 1-like [Jatropha curcas] |
| 2 |
Hb_002007_320 |
0.1054811283 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase ANP1 [Jatropha curcas] |
| 3 |
Hb_000062_090 |
0.1170731423 |
- |
- |
PREDICTED: uncharacterized protein LOC105644743 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000935_040 |
0.1236965829 |
- |
- |
Uncharacterized protein isoform 2 [Theobroma cacao] |
| 5 |
Hb_001904_050 |
0.1241707344 |
- |
- |
alpha-tubulin 1 [Hevea brasiliensis] |
| 6 |
Hb_004158_020 |
0.1242119417 |
- |
- |
Rab1 [Hevea brasiliensis] |
| 7 |
Hb_188281_030 |
0.1309764646 |
- |
- |
PREDICTED: uridine nucleosidase 1 [Jatropha curcas] |
| 8 |
Hb_003813_060 |
0.1311125862 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000023_310 |
0.1325356051 |
- |
- |
PREDICTED: probable folate-biopterin transporter 2 [Populus euphratica] |
| 10 |
Hb_048476_060 |
0.1347460712 |
- |
- |
PREDICTED: CRAL-TRIO domain-containing protein YKL091C-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_000012_080 |
0.135483218 |
- |
- |
PREDICTED: clathrin interactor EPSIN 1 [Jatropha curcas] |
| 12 |
Hb_003602_060 |
0.1439996703 |
- |
- |
PREDICTED: phosphoglycerate mutase-like protein 1 [Jatropha curcas] |
| 13 |
Hb_001369_630 |
0.1442161209 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 3-like [Jatropha curcas] |
| 14 |
Hb_002193_060 |
0.145724195 |
- |
- |
PREDICTED: uncharacterized protein C4orf29 homolog [Pyrus x bretschneideri] |
| 15 |
Hb_000742_020 |
0.1468417467 |
- |
- |
ARF GTPase activator, putative [Ricinus communis] |
| 16 |
Hb_000765_060 |
0.1479388722 |
- |
- |
Alcohol dehydrogenase-like 6 [Glycine soja] |
| 17 |
Hb_093458_010 |
0.1510453684 |
- |
- |
hypothetical protein CISIN_1g0277592mg [Citrus sinensis] |
| 18 |
Hb_001329_070 |
0.1514442562 |
- |
- |
Glycogen synthase kinase-3 beta, putative [Ricinus communis] |
| 19 |
Hb_000110_050 |
0.1520389608 |
- |
- |
- |
| 20 |
Hb_154038_020 |
0.1531181855 |
- |
- |
hypothetical protein JCGZ_05648 [Jatropha curcas] |