| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
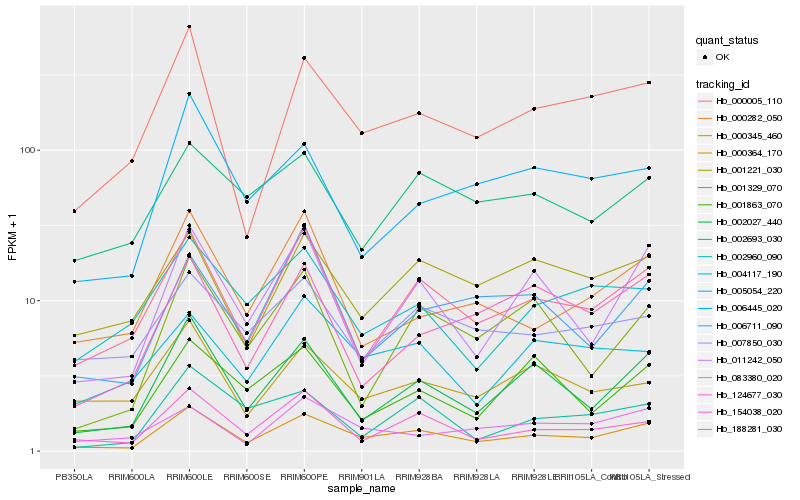
Hb_000345_460 |
0.0 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase ANP1 [Jatropha curcas] |
| 2 |
Hb_188281_030 |
0.1155344015 |
- |
- |
PREDICTED: uridine nucleosidase 1 [Jatropha curcas] |
| 3 |
Hb_002027_440 |
0.1175742803 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_124677_030 |
0.1230424546 |
- |
- |
PREDICTED: uncharacterized protein LOC105650473 [Jatropha curcas] |
| 5 |
Hb_000005_110 |
0.1253272529 |
- |
- |
histone H2B1 [Hevea brasiliensis] |
| 6 |
Hb_011242_050 |
0.1326689802 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2-23 kDa [Jatropha curcas] |
| 7 |
Hb_001221_030 |
0.156249256 |
- |
- |
PREDICTED: adenylate kinase 4 [Jatropha curcas] |
| 8 |
Hb_000282_050 |
0.1609249117 |
- |
- |
calcineurin B, putative [Ricinus communis] |
| 9 |
Hb_000364_170 |
0.1623501194 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase CLF [Jatropha curcas] |
| 10 |
Hb_005054_220 |
0.1626133609 |
- |
- |
PREDICTED: peroxisomal nicotinamide adenine dinucleotide carrier isoform X1 [Jatropha curcas] |
| 11 |
Hb_001863_070 |
0.1631985532 |
- |
- |
Queuine tRNA-ribosyltransferase [Theobroma cacao] |
| 12 |
Hb_002960_090 |
0.1635338958 |
- |
- |
protein with unknown function [Ricinus communis] |
| 13 |
Hb_007850_030 |
0.1646477622 |
- |
- |
PREDICTED: uncharacterized protein LOC105640669 [Jatropha curcas] |
| 14 |
Hb_006445_020 |
0.1653999263 |
- |
- |
hypothetical protein L484_026741 [Morus notabilis] |
| 15 |
Hb_002693_030 |
0.1655451348 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin of 34 kDa [Jatropha curcas] |
| 16 |
Hb_001329_070 |
0.1656913656 |
- |
- |
Glycogen synthase kinase-3 beta, putative [Ricinus communis] |
| 17 |
Hb_004117_190 |
0.1669645643 |
- |
- |
PREDICTED: uncharacterized protein LOC105649109 [Jatropha curcas] |
| 18 |
Hb_006711_090 |
0.1676148531 |
- |
- |
PREDICTED: fatty acid 2-hydroxylase 1-like [Jatropha curcas] |
| 19 |
Hb_083380_020 |
0.1684075467 |
- |
- |
PREDICTED: LAG1 longevity assurance homolog 2 [Jatropha curcas] |
| 20 |
Hb_154038_020 |
0.1688145603 |
- |
- |
hypothetical protein JCGZ_05648 [Jatropha curcas] |