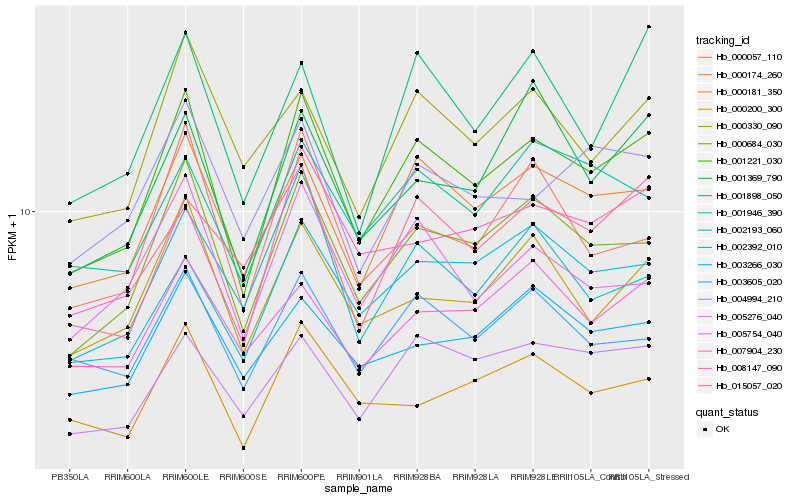
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001221_030 |
0.0 |
- |
- |
PREDICTED: adenylate kinase 4 [Jatropha curcas] |
| 2 |
Hb_000684_030 |
0.0659153447 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 3 |
Hb_000174_260 |
0.074679828 |
- |
- |
PREDICTED: 2-alkenal reductase (NADP(+)-dependent) [Jatropha curcas] |
| 4 |
Hb_005276_040 |
0.0945761906 |
transcription factor |
TF Family: TRAF |
PREDICTED: ARM REPEAT PROTEIN INTERACTING WITH ABF2-like [Jatropha curcas] |
| 5 |
Hb_003605_020 |
0.0973809657 |
- |
- |
exocyst complex component sec6, putative [Ricinus communis] |
| 6 |
Hb_000200_300 |
0.0981868588 |
- |
- |
PREDICTED: uncharacterized protein LOC105636926 [Jatropha curcas] |
| 7 |
Hb_000330_090 |
0.1058874098 |
- |
- |
ornithine carbamoyltransferase, putative [Ricinus communis] |
| 8 |
Hb_003266_030 |
0.1066949709 |
- |
- |
hypothetical protein POPTR_0002s05550g [Populus trichocarpa] |
| 9 |
Hb_004994_210 |
0.1076053248 |
- |
- |
phosphomannomutase family protein [Populus trichocarpa] |
| 10 |
Hb_005754_040 |
0.108032826 |
- |
- |
PREDICTED: uncharacterized protein LOC105636678 isoform X2 [Jatropha curcas] |
| 11 |
Hb_001946_390 |
0.1086835206 |
- |
- |
PREDICTED: NADH-cytochrome b5 reductase-like protein [Jatropha curcas] |
| 12 |
Hb_008147_090 |
0.1090034783 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002392_010 |
0.110278498 |
- |
- |
PREDICTED: apurinic endonuclease-redox protein isoform X4 [Jatropha curcas] |
| 14 |
Hb_001898_050 |
0.110579052 |
- |
- |
PREDICTED: 60S ribosomal protein L3 [Eucalyptus grandis] |
| 15 |
Hb_000057_110 |
0.11069968 |
- |
- |
PREDICTED: kinesin-13A [Jatropha curcas] |
| 16 |
Hb_000181_350 |
0.111571102 |
- |
- |
PREDICTED: uncharacterized protein LOC105111090 [Populus euphratica] |
| 17 |
Hb_007904_230 |
0.1117271846 |
- |
- |
PREDICTED: D-cysteine desulfhydrase 2, mitochondrial [Jatropha curcas] |
| 18 |
Hb_001369_790 |
0.1122455784 |
- |
- |
PREDICTED: clp protease-related protein At4g12060, chloroplastic [Jatropha curcas] |
| 19 |
Hb_015057_020 |
0.1125624295 |
- |
- |
PREDICTED: E3 SUMO-protein ligase MMS21 [Jatropha curcas] |
| 20 |
Hb_002193_060 |
0.116381346 |
- |
- |
PREDICTED: uncharacterized protein C4orf29 homolog [Pyrus x bretschneideri] |