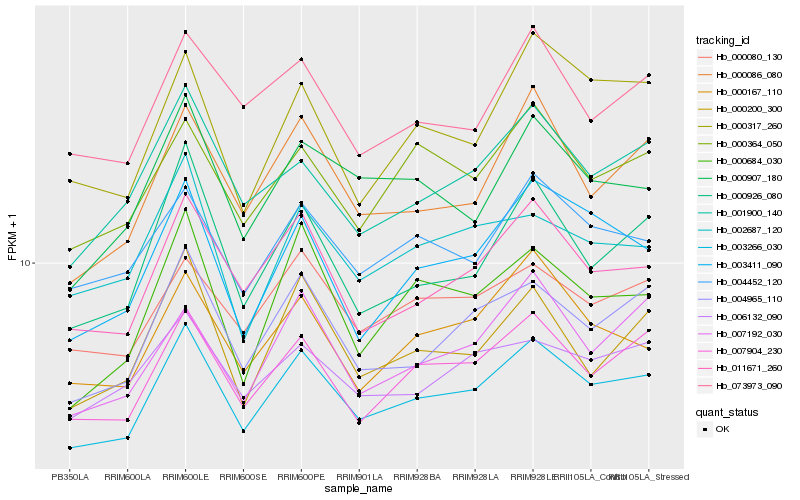
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003266_030 |
0.0 |
- |
- |
hypothetical protein POPTR_0002s05550g [Populus trichocarpa] |
| 2 |
Hb_007904_230 |
0.0589284386 |
- |
- |
PREDICTED: D-cysteine desulfhydrase 2, mitochondrial [Jatropha curcas] |
| 3 |
Hb_011671_260 |
0.0601472589 |
- |
- |
PREDICTED: putative lipase ROG1 isoform X1 [Jatropha curcas] |
| 4 |
Hb_004965_110 |
0.0606587754 |
- |
- |
PREDICTED: folylpolyglutamate synthase [Jatropha curcas] |
| 5 |
Hb_000086_080 |
0.0720479947 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_003411_090 |
0.0730467991 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 4 of pyruvate dehydrogenase complex |
Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase, putative [Ricinus communis] |
| 7 |
Hb_000926_080 |
0.0748541747 |
- |
- |
PREDICTED: paraspeckle component 1 [Jatropha curcas] |
| 8 |
Hb_000080_130 |
0.0751325989 |
- |
- |
PREDICTED: TBC1 domain family member 22B [Jatropha curcas] |
| 9 |
Hb_001900_140 |
0.0752782589 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 4, chloroplastic [Jatropha curcas] |
| 10 |
Hb_004452_120 |
0.0761720607 |
- |
- |
PREDICTED: uncharacterized protein LOC105639574 [Jatropha curcas] |
| 11 |
Hb_000167_110 |
0.076771705 |
- |
- |
glycerol-3-phosphate dehydrogenase, putative [Ricinus communis] |
| 12 |
Hb_000364_050 |
0.0768845599 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX1, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000907_180 |
0.0787312454 |
- |
- |
PREDICTED: uncharacterized protein LOC105641369 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000200_300 |
0.0795578988 |
- |
- |
PREDICTED: uncharacterized protein LOC105636926 [Jatropha curcas] |
| 15 |
Hb_000684_030 |
0.0801196972 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 16 |
Hb_073973_090 |
0.0813337615 |
- |
- |
Aminotransferase ybdL, putative [Ricinus communis] |
| 17 |
Hb_002687_120 |
0.0836049243 |
- |
- |
PREDICTED: lipoyl synthase, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000317_260 |
0.0838440038 |
- |
- |
unknown [Populus trichocarpa] |
| 19 |
Hb_006132_090 |
0.0838834543 |
- |
- |
PREDICTED: origin of replication complex subunit 5 isoform X1 [Jatropha curcas] |
| 20 |
Hb_007192_030 |
0.0849673633 |
- |
- |
PREDICTED: manganese-dependent ADP-ribose/CDP-alcohol diphosphatase [Jatropha curcas] |