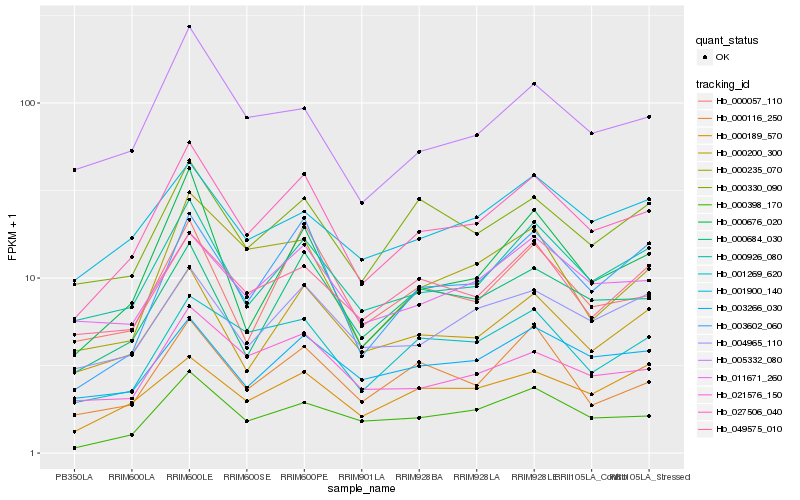
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_027506_040 |
0.0 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit beta-3, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000926_080 |
0.0802219893 |
- |
- |
PREDICTED: paraspeckle component 1 [Jatropha curcas] |
| 3 |
Hb_000200_300 |
0.0942055695 |
- |
- |
PREDICTED: uncharacterized protein LOC105636926 [Jatropha curcas] |
| 4 |
Hb_011671_260 |
0.0973289369 |
- |
- |
PREDICTED: putative lipase ROG1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000398_170 |
0.0974140825 |
- |
- |
PREDICTED: tobamovirus multiplication protein 1-like [Jatropha curcas] |
| 6 |
Hb_003602_060 |
0.0978693023 |
- |
- |
PREDICTED: phosphoglycerate mutase-like protein 1 [Jatropha curcas] |
| 7 |
Hb_000684_030 |
0.0986297067 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 8 |
Hb_001269_620 |
0.0988130909 |
- |
- |
PREDICTED: magnesium transporter MRS2-1 isoform X1 [Populus euphratica] |
| 9 |
Hb_021576_150 |
0.1009335979 |
- |
- |
PREDICTED: formin-like protein 18 [Jatropha curcas] |
| 10 |
Hb_004965_110 |
0.1011969188 |
- |
- |
PREDICTED: folylpolyglutamate synthase [Jatropha curcas] |
| 11 |
Hb_003266_030 |
0.1014538079 |
- |
- |
hypothetical protein POPTR_0002s05550g [Populus trichocarpa] |
| 12 |
Hb_000057_110 |
0.1043604477 |
- |
- |
PREDICTED: kinesin-13A [Jatropha curcas] |
| 13 |
Hb_000330_090 |
0.1052152677 |
- |
- |
ornithine carbamoyltransferase, putative [Ricinus communis] |
| 14 |
Hb_000676_020 |
0.1066942705 |
- |
- |
PREDICTED: uncharacterized protein LOC105645386 [Jatropha curcas] |
| 15 |
Hb_000235_070 |
0.1068986544 |
- |
- |
PREDICTED: VIN3-like protein 2 [Jatropha curcas] |
| 16 |
Hb_000189_570 |
0.1086992877 |
- |
- |
PREDICTED: uncharacterized protein LOC105647652 [Jatropha curcas] |
| 17 |
Hb_000116_250 |
0.1089273944 |
- |
- |
PREDICTED: DNA mismatch repair protein MLH3 [Jatropha curcas] |
| 18 |
Hb_049575_010 |
0.1091285143 |
- |
- |
hypothetical protein POPTR_0001s15330g [Populus trichocarpa] |
| 19 |
Hb_005332_080 |
0.1097135841 |
- |
- |
PREDICTED: succinate dehydrogenase [ubiquinone] iron-sulfur subunit 2, mitochondrial [Jatropha curcas] |
| 20 |
Hb_001900_140 |
0.1097811639 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 4, chloroplastic [Jatropha curcas] |