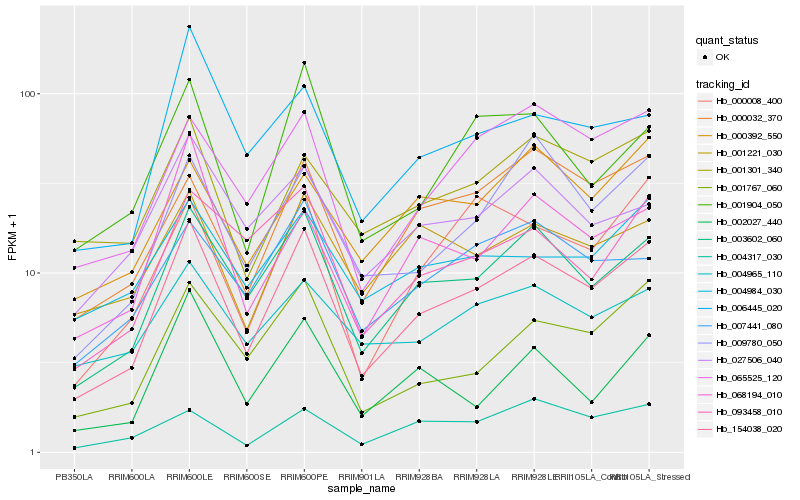
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_154038_020 |
0.0 |
- |
- |
hypothetical protein JCGZ_05648 [Jatropha curcas] |
| 2 |
Hb_003602_060 |
0.0804204448 |
- |
- |
PREDICTED: phosphoglycerate mutase-like protein 1 [Jatropha curcas] |
| 3 |
Hb_001767_060 |
0.1136762754 |
- |
- |
PREDICTED: sulfate transporter 4.1, chloroplastic-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_065525_120 |
0.1183923267 |
- |
- |
dtdp-glucose 4-6-dehydratase, putative [Ricinus communis] |
| 5 |
Hb_009780_050 |
0.1281198828 |
- |
- |
PREDICTED: glutathione S-transferase DHAR3, chloroplastic [Jatropha curcas] |
| 6 |
Hb_001904_050 |
0.1304290396 |
- |
- |
alpha-tubulin 1 [Hevea brasiliensis] |
| 7 |
Hb_093458_010 |
0.13052161 |
- |
- |
hypothetical protein CISIN_1g0277592mg [Citrus sinensis] |
| 8 |
Hb_000032_370 |
0.1310624898 |
- |
- |
PREDICTED: nucleoside diphosphate kinase III, chloroplastic/mitochondrial-like [Citrus sinensis] |
| 9 |
Hb_000008_400 |
0.1341214753 |
- |
- |
PREDICTED: classical arabinogalactan protein 10-like [Nicotiana tomentosiformis] |
| 10 |
Hb_001301_340 |
0.1342795079 |
- |
- |
endoribonuclease L-PSP family protein [Populus trichocarpa] |
| 11 |
Hb_007441_080 |
0.1361939337 |
- |
- |
PREDICTED: ELMO domain-containing protein C-like [Jatropha curcas] |
| 12 |
Hb_068194_010 |
0.14054652 |
- |
- |
hypothetical protein POPTR_0010s23740g [Populus trichocarpa] |
| 13 |
Hb_004317_030 |
0.1406876162 |
- |
- |
PREDICTED: protein CHROMOSOME TRANSMISSION FIDELITY 7 [Jatropha curcas] |
| 14 |
Hb_006445_020 |
0.1411363465 |
- |
- |
hypothetical protein L484_026741 [Morus notabilis] |
| 15 |
Hb_001221_030 |
0.1422090878 |
- |
- |
PREDICTED: adenylate kinase 4 [Jatropha curcas] |
| 16 |
Hb_027506_040 |
0.1429519819 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit beta-3, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000392_550 |
0.1441535179 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 6, chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_002027_440 |
0.1460861129 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_004984_030 |
0.1462016754 |
- |
- |
protein with unknown function [Ricinus communis] |
| 20 |
Hb_004965_110 |
0.1462779219 |
- |
- |
PREDICTED: folylpolyglutamate synthase [Jatropha curcas] |