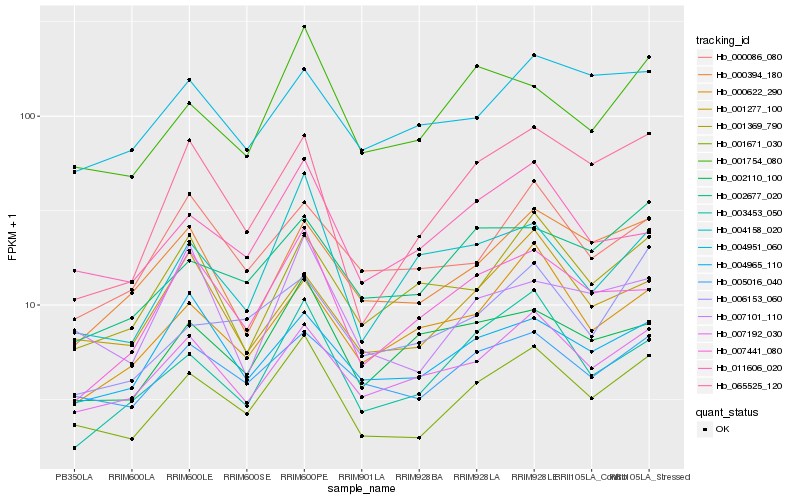
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001671_030 |
0.0 |
- |
- |
PREDICTED: calcium-transporting ATPase, endoplasmic reticulum-type [Jatropha curcas] |
| 2 |
Hb_005016_040 |
0.1033806992 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor-like protein DPB isoform X2 [Jatropha curcas] |
| 3 |
Hb_007192_030 |
0.1037517853 |
- |
- |
PREDICTED: manganese-dependent ADP-ribose/CDP-alcohol diphosphatase [Jatropha curcas] |
| 4 |
Hb_001754_080 |
0.1143063766 |
- |
- |
PREDICTED: protein RALF-like 33 [Jatropha curcas] |
| 5 |
Hb_011606_020 |
0.1165840718 |
- |
- |
hypothetical protein CISIN_1g023687mg [Citrus sinensis] |
| 6 |
Hb_065525_120 |
0.1207745803 |
- |
- |
dtdp-glucose 4-6-dehydratase, putative [Ricinus communis] |
| 7 |
Hb_002677_020 |
0.1209008097 |
- |
- |
PREDICTED: uncharacterized protein LOC105642265 [Jatropha curcas] |
| 8 |
Hb_003453_050 |
0.1215781242 |
- |
- |
PREDICTED: calcium-binding mitochondrial carrier protein SCaMC-3-like [Jatropha curcas] |
| 9 |
Hb_000394_180 |
0.1240055166 |
- |
- |
PREDICTED: uncharacterized protein LOC105636995 [Jatropha curcas] |
| 10 |
Hb_007441_080 |
0.1267815086 |
- |
- |
PREDICTED: ELMO domain-containing protein C-like [Jatropha curcas] |
| 11 |
Hb_002110_100 |
0.127280479 |
- |
- |
PREDICTED: pyridoxine/pyridoxamine 5'-phosphate oxidase 1, chloroplastic [Jatropha curcas] |
| 12 |
Hb_001277_100 |
0.1288196252 |
- |
- |
PREDICTED: 2-methyl-6-phytyl-1,4-hydroquinone methyltransferase, chloroplastic-like isoform X1 [Gossypium raimondii] |
| 13 |
Hb_007101_110 |
0.1289349849 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 7 [Jatropha curcas] |
| 14 |
Hb_004965_110 |
0.1300766078 |
- |
- |
PREDICTED: folylpolyglutamate synthase [Jatropha curcas] |
| 15 |
Hb_004158_020 |
0.1308094222 |
- |
- |
Rab1 [Hevea brasiliensis] |
| 16 |
Hb_000086_080 |
0.131631316 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000622_290 |
0.1319832631 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g61590 [Jatropha curcas] |
| 18 |
Hb_006153_060 |
0.1326345163 |
- |
- |
catalytic, putative [Ricinus communis] |
| 19 |
Hb_001369_790 |
0.1335149362 |
- |
- |
PREDICTED: clp protease-related protein At4g12060, chloroplastic [Jatropha curcas] |
| 20 |
Hb_004951_060 |
0.1340811857 |
- |
- |
PREDICTED: uncharacterized protein LOC105634399 [Jatropha curcas] |