| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
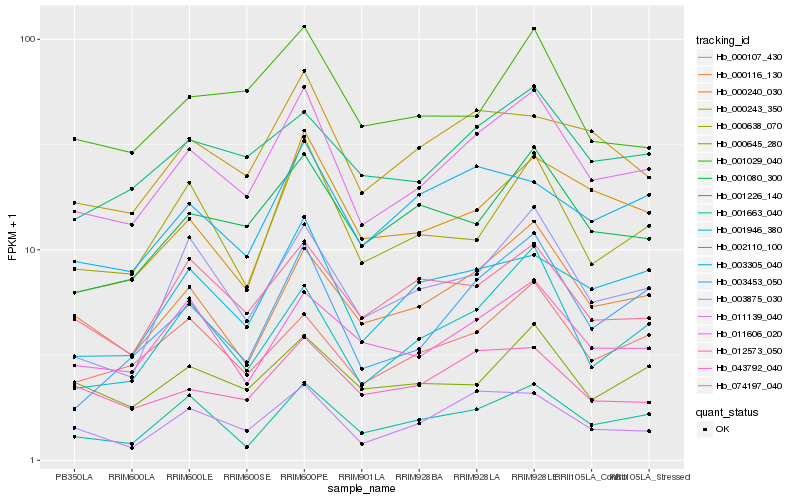
Hb_011606_020 |
0.0 |
- |
- |
hypothetical protein CISIN_1g023687mg [Citrus sinensis] |
| 2 |
Hb_000116_130 |
0.0885162541 |
- |
- |
PREDICTED: aspartic proteinase Asp1 [Jatropha curcas] |
| 3 |
Hb_000107_430 |
0.0956962769 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 1 [Jatropha curcas] |
| 4 |
Hb_001226_140 |
0.0975013437 |
- |
- |
PREDICTED: protein phosphatase 2C 77-like [Jatropha curcas] |
| 5 |
Hb_001080_300 |
0.0994503228 |
- |
- |
PREDICTED: F-box protein SKIP17-like [Jatropha curcas] |
| 6 |
Hb_002110_100 |
0.1014289366 |
- |
- |
PREDICTED: pyridoxine/pyridoxamine 5'-phosphate oxidase 1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_011139_040 |
0.1016369277 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor SPATULA-like isoform X2 [Jatropha curcas] |
| 8 |
Hb_000638_070 |
0.1019794978 |
- |
- |
PREDICTED: protein trichome birefringence-like 14 [Jatropha curcas] |
| 9 |
Hb_001946_380 |
0.1022261726 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Jatropha curcas] |
| 10 |
Hb_003875_030 |
0.1036138427 |
- |
- |
PREDICTED: probable plastidic glucose transporter 1 [Jatropha curcas] |
| 11 |
Hb_000243_350 |
0.1037782803 |
- |
- |
PREDICTED: V-type proton ATPase 16 kDa proteolipid subunit [Tarenaya hassleriana] |
| 12 |
Hb_012573_050 |
0.1049252049 |
- |
- |
choline/ethanolamine kinase family protein [Populus trichocarpa] |
| 13 |
Hb_074197_040 |
0.1084645671 |
- |
- |
PREDICTED: SNARE-interacting protein KEULE [Jatropha curcas] |
| 14 |
Hb_000645_280 |
0.1085201669 |
- |
- |
PREDICTED: AMSH-like ubiquitin thioesterase 3 [Jatropha curcas] |
| 15 |
Hb_043792_040 |
0.1085228792 |
- |
- |
hypothetical protein, partial [Rosa rugosa] |
| 16 |
Hb_003453_050 |
0.1094779534 |
- |
- |
PREDICTED: calcium-binding mitochondrial carrier protein SCaMC-3-like [Jatropha curcas] |
| 17 |
Hb_001663_040 |
0.1096465571 |
- |
- |
hypothetical protein B456_007G162000 [Gossypium raimondii] |
| 18 |
Hb_000240_030 |
0.111227565 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_003305_040 |
0.1126813235 |
- |
- |
AP47/50p mRNA family protein [Populus trichocarpa] |
| 20 |
Hb_001029_040 |
0.113791233 |
- |
- |
PREDICTED: uncharacterized protein LOC105641058 [Jatropha curcas] |