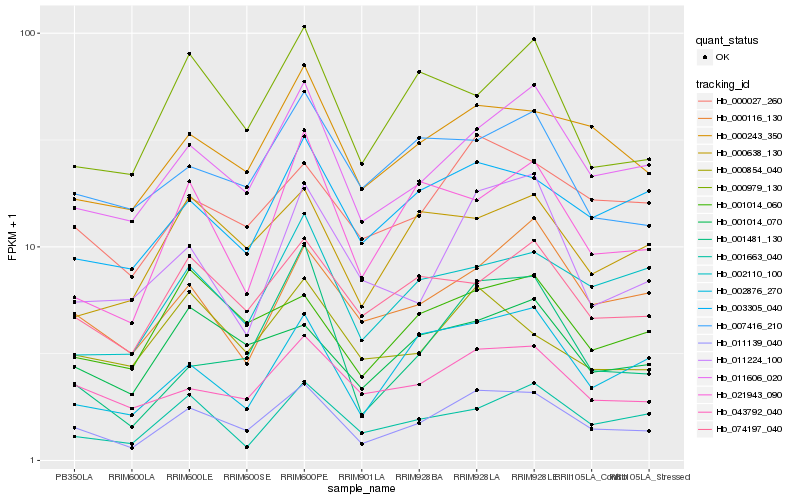
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011139_040 |
0.0 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor SPATULA-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_011606_020 |
0.1016369277 |
- |
- |
hypothetical protein CISIN_1g023687mg [Citrus sinensis] |
| 3 |
Hb_043792_040 |
0.1158306577 |
- |
- |
hypothetical protein, partial [Rosa rugosa] |
| 4 |
Hb_000027_260 |
0.1192192233 |
- |
- |
PREDICTED: chaperone protein dnaJ 15 [Jatropha curcas] |
| 5 |
Hb_000243_350 |
0.1194713666 |
- |
- |
PREDICTED: V-type proton ATPase 16 kDa proteolipid subunit [Tarenaya hassleriana] |
| 6 |
Hb_074197_040 |
0.1287842469 |
- |
- |
PREDICTED: SNARE-interacting protein KEULE [Jatropha curcas] |
| 7 |
Hb_001014_070 |
0.1309892468 |
- |
- |
PREDICTED: uncharacterized protein LOC105647305 [Jatropha curcas] |
| 8 |
Hb_002876_270 |
0.1310456886 |
- |
- |
PREDICTED: probable magnesium transporter NIPA4 isoform X1 [Jatropha curcas] |
| 9 |
Hb_003305_040 |
0.1316791239 |
- |
- |
AP47/50p mRNA family protein [Populus trichocarpa] |
| 10 |
Hb_001014_060 |
0.1351275838 |
transcription factor |
TF Family: SET |
histone-lysine n-methyltransferase, suvh, putative [Ricinus communis] |
| 11 |
Hb_000116_130 |
0.1357947645 |
- |
- |
PREDICTED: aspartic proteinase Asp1 [Jatropha curcas] |
| 12 |
Hb_011224_100 |
0.1377847474 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_021943_090 |
0.1389162834 |
- |
- |
PREDICTED: probable methylenetetrahydrofolate reductase [Jatropha curcas] |
| 14 |
Hb_002110_100 |
0.1399724279 |
- |
- |
PREDICTED: pyridoxine/pyridoxamine 5'-phosphate oxidase 1, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000979_130 |
0.1406716679 |
- |
- |
PREDICTED: phosphoglucomutase, cytoplasmic [Jatropha curcas] |
| 16 |
Hb_000854_040 |
0.1412437236 |
- |
- |
PREDICTED: GPI mannosyltransferase 3 [Jatropha curcas] |
| 17 |
Hb_007416_210 |
0.142110722 |
- |
- |
PREDICTED: coatomer subunit beta-1 [Jatropha curcas] |
| 18 |
Hb_001481_130 |
0.1422863636 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001663_040 |
0.1423630699 |
- |
- |
hypothetical protein B456_007G162000 [Gossypium raimondii] |
| 20 |
Hb_000638_130 |
0.1426507979 |
- |
- |
PREDICTED: SAL1 phosphatase-like [Jatropha curcas] |