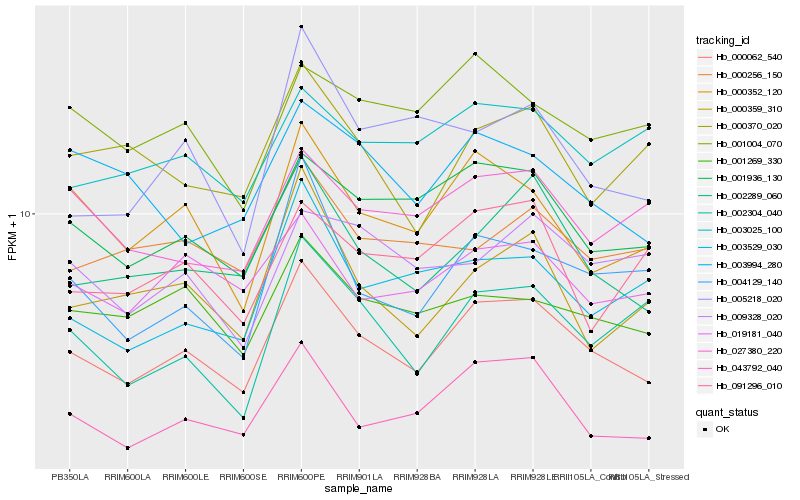
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000062_540 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002289_060 |
0.0730800439 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_043792_040 |
0.0731578363 |
- |
- |
hypothetical protein, partial [Rosa rugosa] |
| 4 |
Hb_001936_130 |
0.0774351296 |
- |
- |
PREDICTED: signal recognition particle receptor subunit beta-like [Jatropha curcas] |
| 5 |
Hb_000352_120 |
0.0839992683 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105630380 [Jatropha curcas] |
| 6 |
Hb_004129_140 |
0.090320112 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At1g06470 [Jatropha curcas] |
| 7 |
Hb_019181_040 |
0.0910017968 |
- |
- |
component of oligomeric golgi complex, putative [Ricinus communis] |
| 8 |
Hb_001269_330 |
0.0914752885 |
- |
- |
PREDICTED: uncharacterized protein LOC105645377 [Jatropha curcas] |
| 9 |
Hb_000359_310 |
0.0977483359 |
- |
- |
hypothetical protein JCGZ_03934 [Jatropha curcas] |
| 10 |
Hb_003529_030 |
0.0990351227 |
- |
- |
Non-imprinted in Prader-Willi/Angelman syndrome region protein, putative [Ricinus communis] |
| 11 |
Hb_027380_220 |
0.0997335572 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 20 [Jatropha curcas] |
| 12 |
Hb_002304_040 |
0.1011904534 |
- |
- |
PREDICTED: uncharacterized protein LOC105649623 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000256_150 |
0.1014876702 |
- |
- |
PREDICTED: trafficking protein particle complex II-specific subunit 120 homolog [Jatropha curcas] |
| 14 |
Hb_003994_280 |
0.1019253106 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g16860-like [Jatropha curcas] |
| 15 |
Hb_003025_100 |
0.1021396167 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 5 isoform X2 [Jatropha curcas] |
| 16 |
Hb_001004_070 |
0.1023038399 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000370_020 |
0.1045896599 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_091296_010 |
0.1079777863 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 19 |
Hb_005218_020 |
0.1082613287 |
- |
- |
Uncharacterized protein isoform 3 [Theobroma cacao] |
| 20 |
Hb_009328_020 |
0.1086470799 |
- |
- |
conserved hypothetical protein [Ricinus communis] |