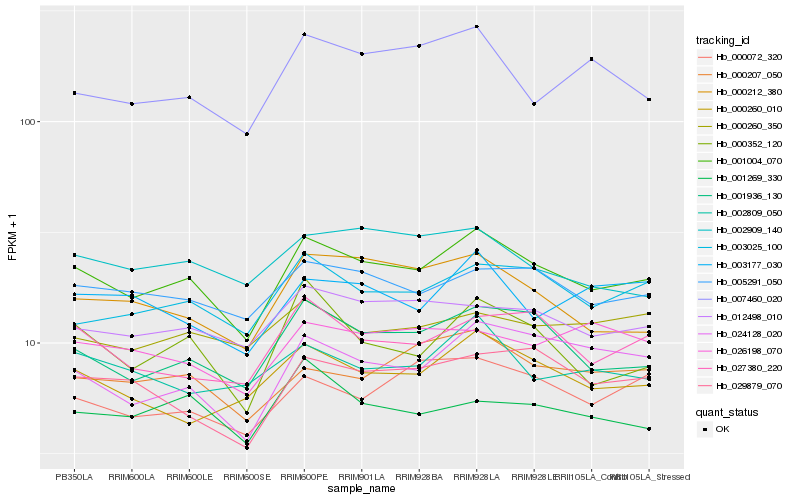
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001004_070 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001936_130 |
0.0611631631 |
- |
- |
PREDICTED: signal recognition particle receptor subunit beta-like [Jatropha curcas] |
| 3 |
Hb_005291_050 |
0.0685743855 |
- |
- |
PREDICTED: mannan endo-1,4-beta-mannosidase 6-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_003025_100 |
0.0722410395 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 5 isoform X2 [Jatropha curcas] |
| 5 |
Hb_012498_010 |
0.0735547292 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM44-2 [Jatropha curcas] |
| 6 |
Hb_027380_220 |
0.0751145504 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 20 [Jatropha curcas] |
| 7 |
Hb_024128_020 |
0.0777632811 |
- |
- |
PREDICTED: vacuolar cation/proton exchanger 5-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_000260_010 |
0.0782097991 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 1-like [Jatropha curcas] |
| 9 |
Hb_000207_050 |
0.0788330575 |
- |
- |
PREDICTED: uncharacterized protein LOC105634369 isoform X1 [Jatropha curcas] |
| 10 |
Hb_029879_070 |
0.0804268692 |
- |
- |
PREDICTED: phospholipase A I [Jatropha curcas] |
| 11 |
Hb_000352_120 |
0.0804305597 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105630380 [Jatropha curcas] |
| 12 |
Hb_000212_380 |
0.0804477549 |
- |
- |
Aspartic proteinase Asp1 precursor, putative [Ricinus communis] |
| 13 |
Hb_000072_320 |
0.0823188001 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RLIM-like [Jatropha curcas] |
| 14 |
Hb_002809_050 |
0.0849216891 |
- |
- |
PREDICTED: protein FIZZY-RELATED 2 [Jatropha curcas] |
| 15 |
Hb_002909_140 |
0.0859419594 |
- |
- |
PREDICTED: D-xylose-proton symporter-like 2 [Jatropha curcas] |
| 16 |
Hb_001269_330 |
0.086641743 |
- |
- |
PREDICTED: uncharacterized protein LOC105645377 [Jatropha curcas] |
| 17 |
Hb_003177_030 |
0.0871791946 |
- |
- |
PREDICTED: uncharacterized protein LOC105635357 [Jatropha curcas] |
| 18 |
Hb_000260_350 |
0.0881717029 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_026198_070 |
0.0888555581 |
- |
- |
PREDICTED: uncharacterized membrane protein At4g09580 [Jatropha curcas] |
| 20 |
Hb_007460_020 |
0.0889401336 |
- |
- |
PREDICTED: guanosine nucleotide diphosphate dissociation inhibitor 1 [Jatropha curcas] |