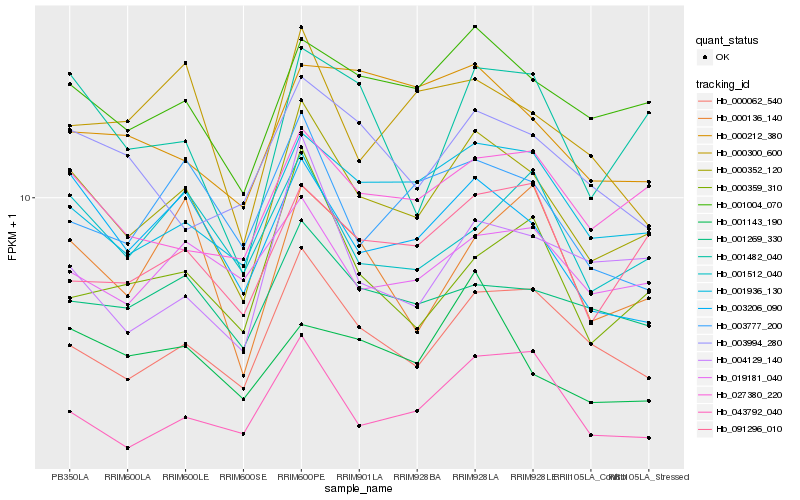
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000352_120 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105630380 [Jatropha curcas] |
| 2 |
Hb_001004_070 |
0.0804305597 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000062_540 |
0.0839992683 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001936_130 |
0.0857050016 |
- |
- |
PREDICTED: signal recognition particle receptor subunit beta-like [Jatropha curcas] |
| 5 |
Hb_003206_090 |
0.0879594894 |
- |
- |
PREDICTED: protein trichome birefringence-like 5 [Jatropha curcas] |
| 6 |
Hb_001269_330 |
0.095151773 |
- |
- |
PREDICTED: uncharacterized protein LOC105645377 [Jatropha curcas] |
| 7 |
Hb_027380_220 |
0.0996810806 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 20 [Jatropha curcas] |
| 8 |
Hb_043792_040 |
0.099747855 |
- |
- |
hypothetical protein, partial [Rosa rugosa] |
| 9 |
Hb_001482_040 |
0.0999381113 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_019181_040 |
0.1049541458 |
- |
- |
component of oligomeric golgi complex, putative [Ricinus communis] |
| 11 |
Hb_003777_200 |
0.1051046041 |
- |
- |
PREDICTED: uncharacterized protein LOC105640933 [Jatropha curcas] |
| 12 |
Hb_091296_010 |
0.1056966406 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 13 |
Hb_001512_040 |
0.109441048 |
- |
- |
PREDICTED: probable methyltransferase PMT9 [Jatropha curcas] |
| 14 |
Hb_001143_190 |
0.1110864825 |
- |
- |
PREDICTED: uncharacterized protein LOC105638976 [Jatropha curcas] |
| 15 |
Hb_004129_140 |
0.1117265842 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At1g06470 [Jatropha curcas] |
| 16 |
Hb_003994_280 |
0.1119274019 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g16860-like [Jatropha curcas] |
| 17 |
Hb_000300_600 |
0.1128445229 |
- |
- |
tubulin alpha chain, putative [Ricinus communis] |
| 18 |
Hb_000359_310 |
0.1130070081 |
- |
- |
hypothetical protein JCGZ_03934 [Jatropha curcas] |
| 19 |
Hb_000212_380 |
0.1145216003 |
- |
- |
Aspartic proteinase Asp1 precursor, putative [Ricinus communis] |
| 20 |
Hb_000136_140 |
0.1149835329 |
- |
- |
PREDICTED: uncharacterized protein LOC105648960 [Jatropha curcas] |