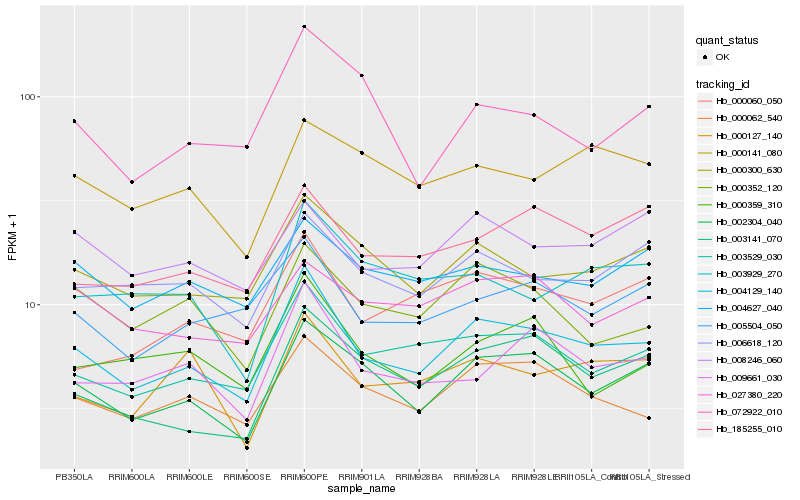
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004129_140 |
0.0 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At1g06470 [Jatropha curcas] |
| 2 |
Hb_002304_040 |
0.079406957 |
- |
- |
PREDICTED: uncharacterized protein LOC105649623 isoform X2 [Jatropha curcas] |
| 3 |
Hb_003529_030 |
0.0838131573 |
- |
- |
Non-imprinted in Prader-Willi/Angelman syndrome region protein, putative [Ricinus communis] |
| 4 |
Hb_000062_540 |
0.090320112 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000300_630 |
0.0925016956 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase-like 7 [Jatropha curcas] |
| 6 |
Hb_009661_030 |
0.1008131877 |
- |
- |
PREDICTED: SPX and EXS domain-containing protein 1-like [Jatropha curcas] |
| 7 |
Hb_005504_050 |
0.1015771547 |
- |
- |
putative protein [Arabidopsis thaliana] |
| 8 |
Hb_006618_120 |
0.1020175877 |
- |
- |
PREDICTED: uncharacterized protein LOC105645969 [Jatropha curcas] |
| 9 |
Hb_003141_070 |
0.1053517034 |
- |
- |
PREDICTED: probable protein phosphatase 2C 51 [Jatropha curcas] |
| 10 |
Hb_004627_040 |
0.1111341964 |
- |
- |
hypothetical protein POPTR_0010s22770g [Populus trichocarpa] |
| 11 |
Hb_000352_120 |
0.1117265842 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105630380 [Jatropha curcas] |
| 12 |
Hb_000127_140 |
0.1124368223 |
- |
- |
transporter-related family protein [Populus trichocarpa] |
| 13 |
Hb_000359_310 |
0.1132756665 |
- |
- |
hypothetical protein JCGZ_03934 [Jatropha curcas] |
| 14 |
Hb_185255_010 |
0.1141061427 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 11-like [Jatropha curcas] |
| 15 |
Hb_008246_060 |
0.1145253728 |
- |
- |
hypothetical protein F383_01577 [Gossypium arboreum] |
| 16 |
Hb_000060_050 |
0.1159724749 |
- |
- |
PREDICTED: uncharacterized protein LOC105649718 [Jatropha curcas] |
| 17 |
Hb_027380_220 |
0.1171686778 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 20 [Jatropha curcas] |
| 18 |
Hb_003929_270 |
0.1175567558 |
- |
- |
S-adenosylmethionine-dependent methyltransferase, putative [Ricinus communis] |
| 19 |
Hb_000141_080 |
0.1177478557 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 8, mitochondrial isoform X1 [Brassica rapa] |
| 20 |
Hb_072922_010 |
0.118106686 |
- |
- |
apolipoprotein d, putative [Ricinus communis] |