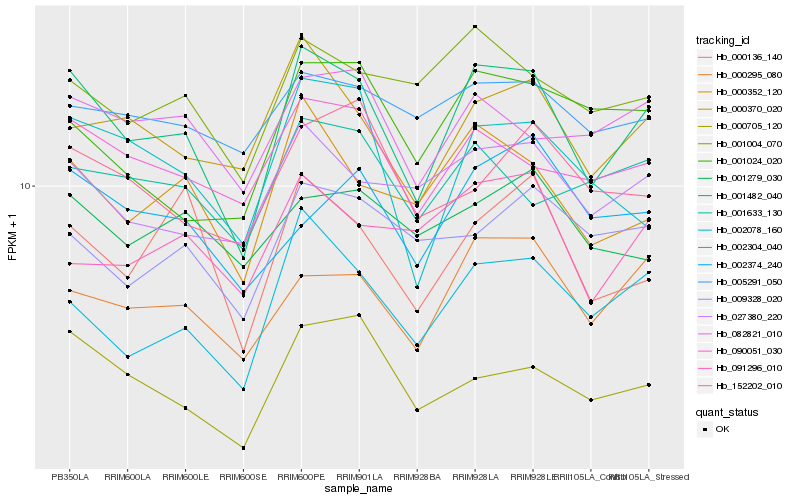
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001482_040 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002304_040 |
0.0993927758 |
- |
- |
PREDICTED: uncharacterized protein LOC105649623 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000295_080 |
0.0999288973 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase [Jatropha curcas] |
| 4 |
Hb_000352_120 |
0.0999381113 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105630380 [Jatropha curcas] |
| 5 |
Hb_000370_020 |
0.1079530004 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_090051_030 |
0.1086089085 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 7 |
Hb_027380_220 |
0.1114488895 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 20 [Jatropha curcas] |
| 8 |
Hb_000705_120 |
0.1121652675 |
- |
- |
glycosyltransferase, putative [Ricinus communis] |
| 9 |
Hb_082821_010 |
0.1144678883 |
- |
- |
PREDICTED: dephospho-CoA kinase-like [Jatropha curcas] |
| 10 |
Hb_002078_160 |
0.1149185916 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 [Jatropha curcas] |
| 11 |
Hb_000136_140 |
0.1153787461 |
- |
- |
PREDICTED: uncharacterized protein LOC105648960 [Jatropha curcas] |
| 12 |
Hb_001004_070 |
0.116135638 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001633_130 |
0.1222435012 |
- |
- |
PREDICTED: uncharacterized protein LOC105630200 [Jatropha curcas] |
| 14 |
Hb_002374_240 |
0.1252128572 |
- |
- |
nucellin, putative [Ricinus communis] |
| 15 |
Hb_009328_020 |
0.1263008087 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001024_020 |
0.1274369541 |
- |
- |
hypothetical protein JCGZ_08989 [Jatropha curcas] |
| 17 |
Hb_152202_010 |
0.1276197757 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 1 [Jatropha curcas] |
| 18 |
Hb_091296_010 |
0.1289005148 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 19 |
Hb_005291_050 |
0.1293001923 |
- |
- |
PREDICTED: mannan endo-1,4-beta-mannosidase 6-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_001279_030 |
0.1299276011 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 57 kDa regulatory subunit B' theta isoform-like [Jatropha curcas] |