| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
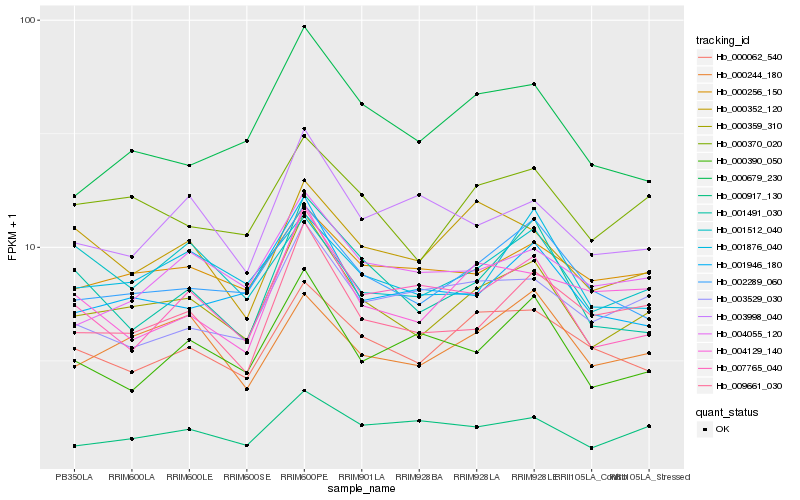
Hb_000359_310 |
0.0 |
- |
- |
hypothetical protein JCGZ_03934 [Jatropha curcas] |
| 2 |
Hb_000370_020 |
0.0823331463 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002289_060 |
0.0892136949 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_009661_030 |
0.0913042417 |
- |
- |
PREDICTED: SPX and EXS domain-containing protein 1-like [Jatropha curcas] |
| 5 |
Hb_001946_180 |
0.0970527465 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000062_540 |
0.0977483359 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000256_150 |
0.1018835885 |
- |
- |
PREDICTED: trafficking protein particle complex II-specific subunit 120 homolog [Jatropha curcas] |
| 8 |
Hb_000244_180 |
0.102281739 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_007765_040 |
0.1026002921 |
- |
- |
Cytosolic enolase isoform 3 [Theobroma cacao] |
| 10 |
Hb_000917_130 |
0.1051686145 |
- |
- |
PREDICTED: uncharacterized protein LOC105635730 [Jatropha curcas] |
| 11 |
Hb_001491_030 |
0.1055217852 |
- |
- |
PREDICTED: uncharacterized protein LOC105649804 isoform X1 [Jatropha curcas] |
| 12 |
Hb_004055_120 |
0.1058494685 |
- |
- |
PREDICTED: uncharacterized protein LOC105631443 [Jatropha curcas] |
| 13 |
Hb_000679_230 |
0.1098909598 |
- |
- |
PREDICTED: serine/threonine-protein kinase PRP4 homolog [Jatropha curcas] |
| 14 |
Hb_001876_040 |
0.110571699 |
- |
- |
PREDICTED: mucin-5B [Jatropha curcas] |
| 15 |
Hb_001512_040 |
0.1128581332 |
- |
- |
PREDICTED: probable methyltransferase PMT9 [Jatropha curcas] |
| 16 |
Hb_000390_050 |
0.1129144063 |
- |
- |
PREDICTED: GPCR-type G protein 1 [Jatropha curcas] |
| 17 |
Hb_000352_120 |
0.1130070081 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105630380 [Jatropha curcas] |
| 18 |
Hb_003529_030 |
0.1130812529 |
- |
- |
Non-imprinted in Prader-Willi/Angelman syndrome region protein, putative [Ricinus communis] |
| 19 |
Hb_004129_140 |
0.1132756665 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At1g06470 [Jatropha curcas] |
| 20 |
Hb_003998_040 |
0.1138075359 |
- |
- |
organic anion transporter, putative [Ricinus communis] |