| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
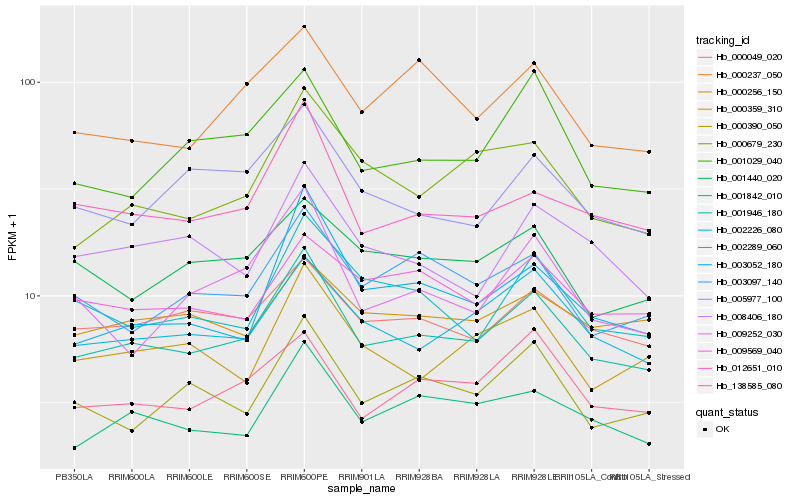
Hb_001946_180 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002289_060 |
0.0768745967 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_012651_010 |
0.0804376654 |
- |
- |
PREDICTED: uncharacterized protein LOC105645131 [Jatropha curcas] |
| 4 |
Hb_001842_010 |
0.0820463184 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 5 |
Hb_000049_020 |
0.0861542221 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 6 |
Hb_138585_080 |
0.0870001231 |
- |
- |
PREDICTED: uncharacterized protein LOC105648741 [Jatropha curcas] |
| 7 |
Hb_002226_080 |
0.0885162706 |
- |
- |
PREDICTED: uncharacterized protein LOC105641402 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000679_230 |
0.0923786003 |
- |
- |
PREDICTED: serine/threonine-protein kinase PRP4 homolog [Jatropha curcas] |
| 9 |
Hb_003097_140 |
0.0923948404 |
- |
- |
PREDICTED: glycosyltransferase-like KOBITO 1 [Jatropha curcas] |
| 10 |
Hb_003052_180 |
0.093057028 |
- |
- |
Diacylglycerol Cholinephosphotransferase [Ricinus communis] |
| 11 |
Hb_000390_050 |
0.0931876907 |
- |
- |
PREDICTED: GPCR-type G protein 1 [Jatropha curcas] |
| 12 |
Hb_001029_040 |
0.0933389846 |
- |
- |
PREDICTED: uncharacterized protein LOC105641058 [Jatropha curcas] |
| 13 |
Hb_005977_100 |
0.0935923965 |
- |
- |
PREDICTED: tetraspanin-18 [Jatropha curcas] |
| 14 |
Hb_009252_030 |
0.0937451876 |
- |
- |
PREDICTED: CASP-like protein 4A3 [Jatropha curcas] |
| 15 |
Hb_000237_050 |
0.0962212248 |
- |
- |
CP2 [Hevea brasiliensis] |
| 16 |
Hb_008406_180 |
0.0969211828 |
- |
- |
RING/U-box superfamily protein isoform 2 [Theobroma cacao] |
| 17 |
Hb_000359_310 |
0.0970527465 |
- |
- |
hypothetical protein JCGZ_03934 [Jatropha curcas] |
| 18 |
Hb_009569_040 |
0.0980402223 |
- |
- |
PREDICTED: uncharacterized protein LOC105635573 [Jatropha curcas] |
| 19 |
Hb_001440_020 |
0.0990430983 |
- |
- |
exocyst componenet sec8, putative [Ricinus communis] |
| 20 |
Hb_000256_150 |
0.0992987537 |
- |
- |
PREDICTED: trafficking protein particle complex II-specific subunit 120 homolog [Jatropha curcas] |