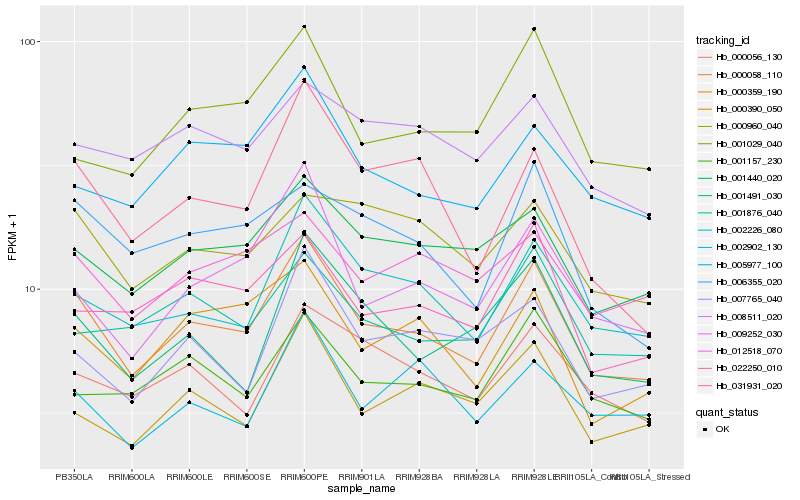
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000058_110 |
0.0 |
- |
- |
PREDICTED: putative BPI/LBP family protein At1g04970 isoform X1 [Jatropha curcas] |
| 2 |
Hb_002226_080 |
0.0857165296 |
- |
- |
PREDICTED: uncharacterized protein LOC105641402 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001440_020 |
0.0932978229 |
- |
- |
exocyst componenet sec8, putative [Ricinus communis] |
| 4 |
Hb_009252_030 |
0.0980537102 |
- |
- |
PREDICTED: CASP-like protein 4A3 [Jatropha curcas] |
| 5 |
Hb_001491_030 |
0.1006919237 |
- |
- |
PREDICTED: uncharacterized protein LOC105649804 isoform X1 [Jatropha curcas] |
| 6 |
Hb_005977_100 |
0.100794914 |
- |
- |
PREDICTED: tetraspanin-18 [Jatropha curcas] |
| 7 |
Hb_000359_190 |
0.1010001839 |
- |
- |
PREDICTED: protein VACUOLELESS1 [Nicotiana sylvestris] |
| 8 |
Hb_000390_050 |
0.101365053 |
- |
- |
PREDICTED: GPCR-type G protein 1 [Jatropha curcas] |
| 9 |
Hb_022250_010 |
0.1020032882 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX10L [Jatropha curcas] |
| 10 |
Hb_000056_130 |
0.1021973973 |
- |
- |
Sucrose Transporter 2C [Hevea brasiliensis subsp. brasiliensis] |
| 11 |
Hb_001157_230 |
0.1038315415 |
- |
- |
Protein AFR, putative [Ricinus communis] |
| 12 |
Hb_006355_020 |
0.1053835685 |
- |
- |
PREDICTED: CLIP-associated protein [Jatropha curcas] |
| 13 |
Hb_007765_040 |
0.1060506509 |
- |
- |
Cytosolic enolase isoform 3 [Theobroma cacao] |
| 14 |
Hb_001876_040 |
0.1070888381 |
- |
- |
PREDICTED: mucin-5B [Jatropha curcas] |
| 15 |
Hb_012518_070 |
0.1079772679 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 9 isoform X1 [Jatropha curcas] |
| 16 |
Hb_031931_020 |
0.1080894723 |
- |
- |
Minor histocompatibility antigen H13, putative [Ricinus communis] |
| 17 |
Hb_008511_020 |
0.1099227665 |
- |
- |
PREDICTED: coatomer subunit gamma-2 [Jatropha curcas] |
| 18 |
Hb_001029_040 |
0.1112807695 |
- |
- |
PREDICTED: uncharacterized protein LOC105641058 [Jatropha curcas] |
| 19 |
Hb_000960_040 |
0.112364536 |
- |
- |
PREDICTED: ERAD-associated E3 ubiquitin-protein ligase HRD1B-like [Vitis vinifera] |
| 20 |
Hb_002902_130 |
0.1128575137 |
- |
- |
PREDICTED: molybdate-anion transporter [Jatropha curcas] |