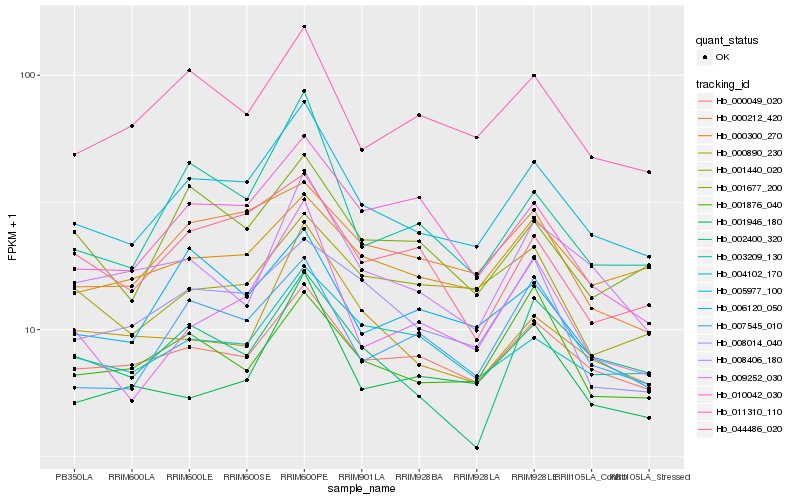
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005977_100 |
0.0 |
- |
- |
PREDICTED: tetraspanin-18 [Jatropha curcas] |
| 2 |
Hb_000049_020 |
0.0752617186 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 3 |
Hb_007545_010 |
0.0773509044 |
- |
- |
HIV-1 rev binding protein, hrbl, putative [Ricinus communis] |
| 4 |
Hb_011310_110 |
0.0783874247 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At5g25400 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000212_420 |
0.0788902782 |
- |
- |
PREDICTED: probable protein phosphatase 2C 33 [Jatropha curcas] |
| 6 |
Hb_003209_130 |
0.0799784935 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 7 |
Hb_004102_170 |
0.0854805525 |
- |
- |
PREDICTED: lys-63-specific deubiquitinase BRCC36-like [Jatropha curcas] |
| 8 |
Hb_001677_200 |
0.0858394552 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000300_270 |
0.0876100462 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 10 |
Hb_044486_020 |
0.088227472 |
- |
- |
CASTOR protein [Glycine max] |
| 11 |
Hb_006120_050 |
0.0893229781 |
- |
- |
PREDICTED: importin subunit alpha-4-like [Jatropha curcas] |
| 12 |
Hb_002400_320 |
0.0900176962 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_008014_040 |
0.0903527039 |
- |
- |
PREDICTED: uncharacterized protein LOC105649721 [Jatropha curcas] |
| 14 |
Hb_000890_230 |
0.0911038862 |
- |
- |
hypothetical protein JCGZ_02013 [Jatropha curcas] |
| 15 |
Hb_009252_030 |
0.0912600985 |
- |
- |
PREDICTED: CASP-like protein 4A3 [Jatropha curcas] |
| 16 |
Hb_008406_180 |
0.0927218542 |
- |
- |
RING/U-box superfamily protein isoform 2 [Theobroma cacao] |
| 17 |
Hb_010042_030 |
0.0929481175 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine diphosphorylase 1 [Jatropha curcas] |
| 18 |
Hb_001946_180 |
0.0935923965 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001876_040 |
0.0936268629 |
- |
- |
PREDICTED: mucin-5B [Jatropha curcas] |
| 20 |
Hb_001440_020 |
0.0941001306 |
- |
- |
exocyst componenet sec8, putative [Ricinus communis] |