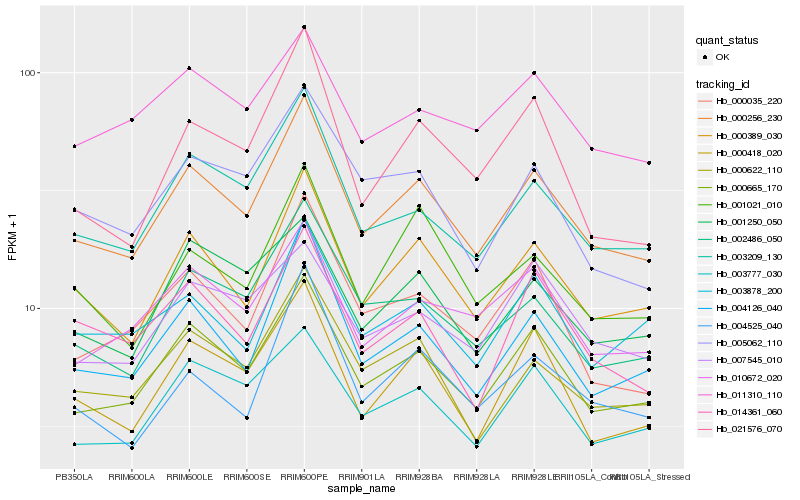
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000256_230 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105637594 [Jatropha curcas] |
| 2 |
Hb_000665_170 |
0.0428158589 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 10 [Jatropha curcas] |
| 3 |
Hb_000389_030 |
0.0484619428 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 1B [Jatropha curcas] |
| 4 |
Hb_003209_130 |
0.0611964532 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 5 |
Hb_004126_040 |
0.0639309252 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000622_110 |
0.0689302956 |
- |
- |
cmp-sialic acid transporter, putative [Ricinus communis] |
| 7 |
Hb_002486_050 |
0.0708524653 |
- |
- |
Multiple inositol polyphosphate phosphatase 1 precursor, putative [Ricinus communis] |
| 8 |
Hb_000418_020 |
0.0719637994 |
- |
- |
PREDICTED: sulfhydryl oxidase 2 isoform X1 [Jatropha curcas] |
| 9 |
Hb_003777_030 |
0.0755638432 |
- |
- |
Exocyst complex component, putative [Ricinus communis] |
| 10 |
Hb_010672_020 |
0.0783976072 |
- |
- |
PREDICTED: ATP synthase gamma chain, chloroplastic-like [Jatropha curcas] |
| 11 |
Hb_000035_220 |
0.0806532421 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 8 [Jatropha curcas] |
| 12 |
Hb_001021_010 |
0.0830831259 |
- |
- |
PREDICTED: bifunctional protein FolD 2 [Jatropha curcas] |
| 13 |
Hb_001250_050 |
0.0831013254 |
- |
- |
PREDICTED: uncharacterized protein LOC105644623 isoform X1 [Jatropha curcas] |
| 14 |
Hb_003878_200 |
0.0842513104 |
- |
- |
PREDICTED: V-type proton ATPase subunit D-like [Jatropha curcas] |
| 15 |
Hb_014361_060 |
0.0846701387 |
- |
- |
PREDICTED: probable prolyl 4-hydroxylase 9 [Jatropha curcas] |
| 16 |
Hb_004525_040 |
0.0848607481 |
- |
- |
PREDICTED: sugar transporter ERD6-like 6 [Jatropha curcas] |
| 17 |
Hb_005062_110 |
0.086410412 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 1 [Jatropha curcas] |
| 18 |
Hb_021576_070 |
0.0870412576 |
- |
- |
hypothetical protein PRUPE_ppa002736mg [Prunus persica] |
| 19 |
Hb_011310_110 |
0.0880304748 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At5g25400 isoform X1 [Jatropha curcas] |
| 20 |
Hb_007545_010 |
0.0891480924 |
- |
- |
HIV-1 rev binding protein, hrbl, putative [Ricinus communis] |