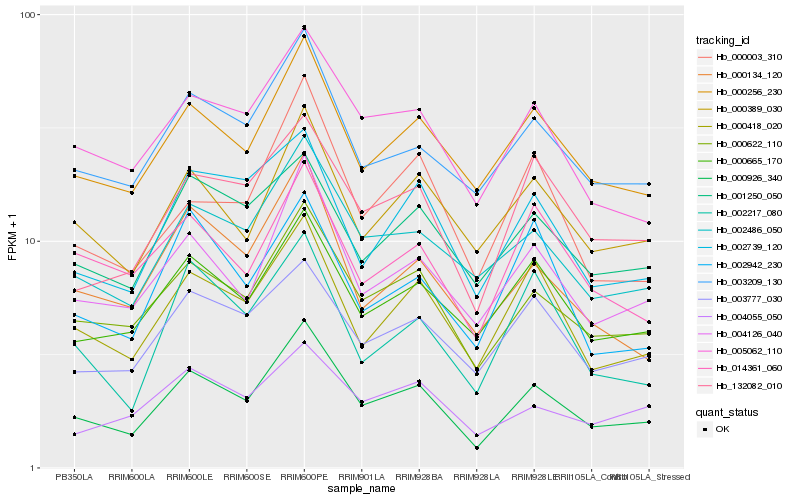
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000926_340 |
0.0 |
- |
- |
PREDICTED: EH domain-containing protein 1 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000622_110 |
0.0761369728 |
- |
- |
cmp-sialic acid transporter, putative [Ricinus communis] |
| 3 |
Hb_004126_040 |
0.0873795319 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000418_020 |
0.0906901438 |
- |
- |
PREDICTED: sulfhydryl oxidase 2 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000256_230 |
0.094174563 |
- |
- |
PREDICTED: uncharacterized protein LOC105637594 [Jatropha curcas] |
| 6 |
Hb_002486_050 |
0.0959178028 |
- |
- |
Multiple inositol polyphosphate phosphatase 1 precursor, putative [Ricinus communis] |
| 7 |
Hb_003209_130 |
0.0975202997 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 8 |
Hb_005062_110 |
0.1000595366 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 1 [Jatropha curcas] |
| 9 |
Hb_132082_010 |
0.1025868903 |
- |
- |
hypothetical protein VITISV_030895 [Vitis vinifera] |
| 10 |
Hb_000389_030 |
0.1028141586 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 1B [Jatropha curcas] |
| 11 |
Hb_000665_170 |
0.1035185969 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 10 [Jatropha curcas] |
| 12 |
Hb_000134_120 |
0.103959073 |
- |
- |
PREDICTED: uncharacterized protein LOC105628669 [Jatropha curcas] |
| 13 |
Hb_000003_310 |
0.1070098905 |
- |
- |
PREDICTED: glycosyltransferase-like KOBITO 1 [Jatropha curcas] |
| 14 |
Hb_002739_120 |
0.1083634635 |
- |
- |
PREDICTED: zinc finger protein ZPR1-like [Jatropha curcas] |
| 15 |
Hb_002217_080 |
0.1119286057 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 16 [Jatropha curcas] |
| 16 |
Hb_003777_030 |
0.1128563684 |
- |
- |
Exocyst complex component, putative [Ricinus communis] |
| 17 |
Hb_001250_050 |
0.1172954931 |
- |
- |
PREDICTED: uncharacterized protein LOC105644623 isoform X1 [Jatropha curcas] |
| 18 |
Hb_014361_060 |
0.1182109431 |
- |
- |
PREDICTED: probable prolyl 4-hydroxylase 9 [Jatropha curcas] |
| 19 |
Hb_004055_050 |
0.1203876759 |
- |
- |
PREDICTED: coatomer subunit zeta-1-like [Jatropha curcas] |
| 20 |
Hb_002942_230 |
0.121711616 |
- |
- |
PREDICTED: long chain base biosynthesis protein 2a [Populus euphratica] |