| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
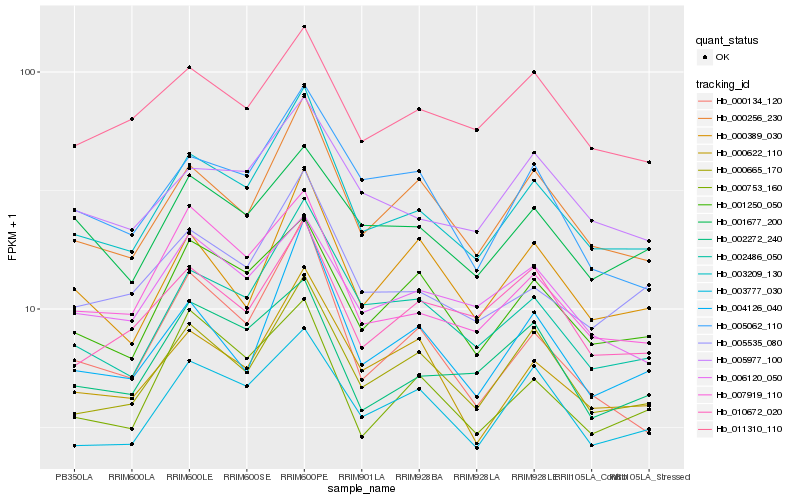
Hb_003209_130 |
0.0 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 2 |
Hb_002486_050 |
0.0604020813 |
- |
- |
Multiple inositol polyphosphate phosphatase 1 precursor, putative [Ricinus communis] |
| 3 |
Hb_000256_230 |
0.0611964532 |
- |
- |
PREDICTED: uncharacterized protein LOC105637594 [Jatropha curcas] |
| 4 |
Hb_000665_170 |
0.0669080053 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 10 [Jatropha curcas] |
| 5 |
Hb_005535_080 |
0.0724239382 |
- |
- |
hypothetical protein CICLE_v10012766mg [Citrus clementina] |
| 6 |
Hb_000622_110 |
0.0779959432 |
- |
- |
cmp-sialic acid transporter, putative [Ricinus communis] |
| 7 |
Hb_004126_040 |
0.078681074 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_005977_100 |
0.0799784935 |
- |
- |
PREDICTED: tetraspanin-18 [Jatropha curcas] |
| 9 |
Hb_003777_030 |
0.0801804869 |
- |
- |
Exocyst complex component, putative [Ricinus communis] |
| 10 |
Hb_007919_110 |
0.0805375194 |
- |
- |
PREDICTED: UDP-galactose transporter 2-like [Jatropha curcas] |
| 11 |
Hb_001250_050 |
0.0820357726 |
- |
- |
PREDICTED: uncharacterized protein LOC105644623 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000134_120 |
0.0870262929 |
- |
- |
PREDICTED: uncharacterized protein LOC105628669 [Jatropha curcas] |
| 13 |
Hb_005062_110 |
0.089405195 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 1 [Jatropha curcas] |
| 14 |
Hb_000389_030 |
0.0922206682 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 1B [Jatropha curcas] |
| 15 |
Hb_010672_020 |
0.0934260249 |
- |
- |
PREDICTED: ATP synthase gamma chain, chloroplastic-like [Jatropha curcas] |
| 16 |
Hb_006120_050 |
0.0941392789 |
- |
- |
PREDICTED: importin subunit alpha-4-like [Jatropha curcas] |
| 17 |
Hb_002272_240 |
0.0941966079 |
- |
- |
catalytic, putative [Ricinus communis] |
| 18 |
Hb_011310_110 |
0.0945320891 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At5g25400 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000753_160 |
0.0948960012 |
- |
- |
PREDICTED: uncharacterized protein LOC105640669 [Jatropha curcas] |
| 20 |
Hb_001677_200 |
0.095609472 |
- |
- |
conserved hypothetical protein [Ricinus communis] |