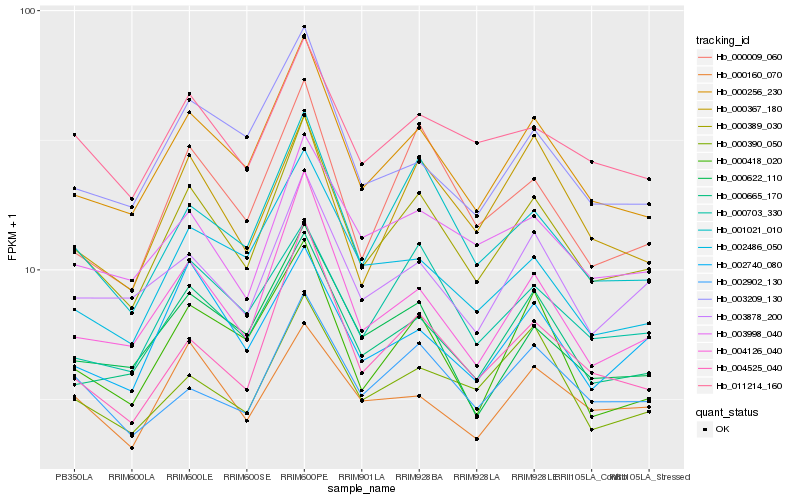
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000389_030 |
0.0 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 1B [Jatropha curcas] |
| 2 |
Hb_000256_230 |
0.0484619428 |
- |
- |
PREDICTED: uncharacterized protein LOC105637594 [Jatropha curcas] |
| 3 |
Hb_001021_010 |
0.0668899095 |
- |
- |
PREDICTED: bifunctional protein FolD 2 [Jatropha curcas] |
| 4 |
Hb_004126_040 |
0.0681857533 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000665_170 |
0.0732641486 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 10 [Jatropha curcas] |
| 6 |
Hb_004525_040 |
0.0772182772 |
- |
- |
PREDICTED: sugar transporter ERD6-like 6 [Jatropha curcas] |
| 7 |
Hb_000009_060 |
0.0781831619 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 1B [Jatropha curcas] |
| 8 |
Hb_003878_200 |
0.0814047147 |
- |
- |
PREDICTED: V-type proton ATPase subunit D-like [Jatropha curcas] |
| 9 |
Hb_011214_160 |
0.0814375977 |
- |
- |
PREDICTED: coatomer subunit delta [Jatropha curcas] |
| 10 |
Hb_003998_040 |
0.08377491 |
- |
- |
organic anion transporter, putative [Ricinus communis] |
| 11 |
Hb_000418_020 |
0.0839723706 |
- |
- |
PREDICTED: sulfhydryl oxidase 2 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000622_110 |
0.0857556644 |
- |
- |
cmp-sialic acid transporter, putative [Ricinus communis] |
| 13 |
Hb_000703_330 |
0.0858941341 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002902_130 |
0.086578787 |
- |
- |
PREDICTED: molybdate-anion transporter [Jatropha curcas] |
| 15 |
Hb_000390_050 |
0.0914444984 |
- |
- |
PREDICTED: GPCR-type G protein 1 [Jatropha curcas] |
| 16 |
Hb_003209_130 |
0.0922206682 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 17 |
Hb_002486_050 |
0.0925596503 |
- |
- |
Multiple inositol polyphosphate phosphatase 1 precursor, putative [Ricinus communis] |
| 18 |
Hb_002740_080 |
0.0928122332 |
- |
- |
flap endonuclease-1, putative [Ricinus communis] |
| 19 |
Hb_000367_180 |
0.0934989433 |
- |
- |
Heparanase-2, putative [Ricinus communis] |
| 20 |
Hb_000160_070 |
0.0942751668 |
- |
- |
PREDICTED: uncharacterized protein LOC105648219 [Jatropha curcas] |