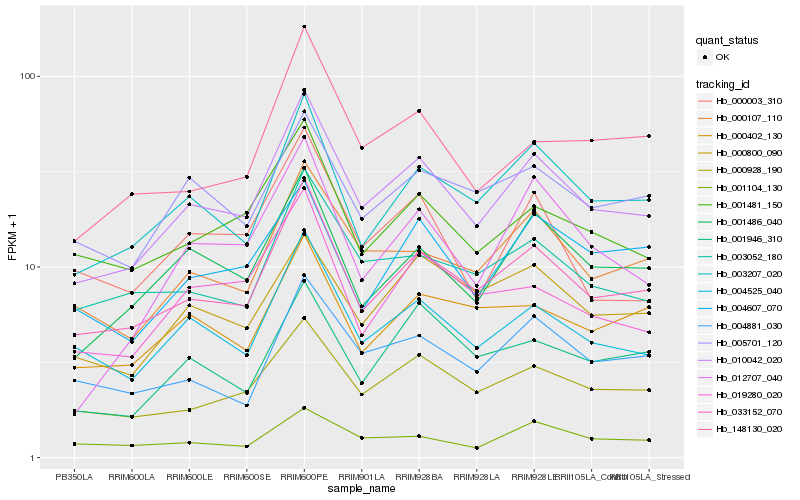
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010042_020 |
0.0 |
- |
- |
UDP-n-acteylglucosamine pyrophosphorylase, putative [Ricinus communis] |
| 2 |
Hb_033152_070 |
0.058749893 |
- |
- |
N-acetyl-gamma-glutamyl-phosphate reductase, putative [Ricinus communis] |
| 3 |
Hb_003207_020 |
0.0822482597 |
- |
- |
PREDICTED: bifunctional dTDP-4-dehydrorhamnose 3,5-epimerase/dTDP-4-dehydrorhamnose reductase [Jatropha curcas] |
| 4 |
Hb_000928_190 |
0.0858667686 |
- |
- |
PREDICTED: putative acyl-activating enzyme 19 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001486_040 |
0.0869593331 |
- |
- |
PREDICTED: uncharacterized protein LOC105632624 isoform X1 [Jatropha curcas] |
| 6 |
Hb_004525_040 |
0.0873570834 |
- |
- |
PREDICTED: sugar transporter ERD6-like 6 [Jatropha curcas] |
| 7 |
Hb_000107_110 |
0.0884693515 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 8 |
Hb_001481_150 |
0.1009542175 |
- |
- |
hypothetical protein POPTR_0001s15440g [Populus trichocarpa] |
| 9 |
Hb_003052_180 |
0.1024866931 |
- |
- |
Diacylglycerol Cholinephosphotransferase [Ricinus communis] |
| 10 |
Hb_005701_120 |
0.106579913 |
- |
- |
PREDICTED: V-type proton ATPase 16 kDa proteolipid subunit [Cucumis sativus] |
| 11 |
Hb_004881_030 |
0.1077410176 |
- |
- |
PREDICTED: uncharacterized protein LOC105629532 isoform X1 [Jatropha curcas] |
| 12 |
Hb_148130_020 |
0.108591951 |
- |
- |
- |
| 13 |
Hb_000800_090 |
0.1095908093 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 1 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 1 of pyruvate dehydrogenase complex, mitochondrial [Jatropha curcas] |
| 14 |
Hb_019280_020 |
0.1113202279 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK14 isoform X1 [Jatropha curcas] |
| 15 |
Hb_004607_070 |
0.1133642316 |
- |
- |
integral membrane protein, putative [Ricinus communis] |
| 16 |
Hb_012707_040 |
0.1142768148 |
- |
- |
PREDICTED: probable inactive purple acid phosphatase 29 [Jatropha curcas] |
| 17 |
Hb_001946_310 |
0.1150260932 |
- |
- |
PREDICTED: probable ethanolamine kinase isoform X2 [Jatropha curcas] |
| 18 |
Hb_000402_130 |
0.1159765568 |
- |
- |
PREDICTED: AP-1 complex subunit mu-2 [Jatropha curcas] |
| 19 |
Hb_000003_310 |
0.1172608865 |
- |
- |
PREDICTED: glycosyltransferase-like KOBITO 1 [Jatropha curcas] |
| 20 |
Hb_001104_130 |
0.1173453966 |
- |
- |
C-14 sterol reductase, putative [Ricinus communis] |