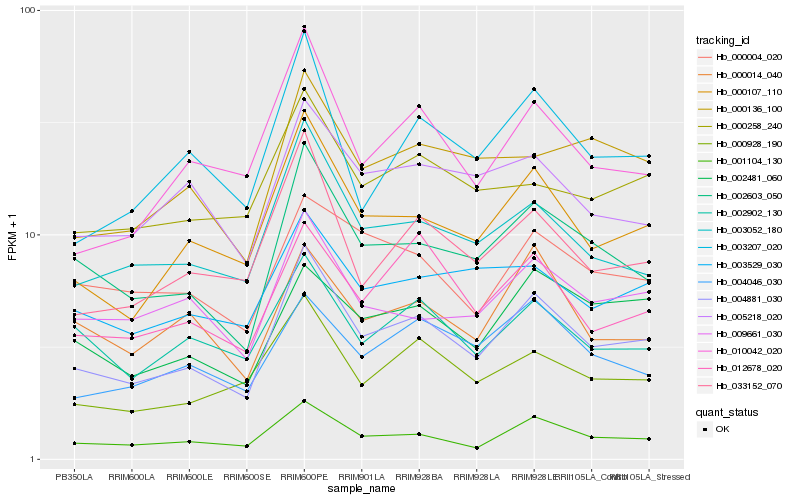
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004881_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105629532 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001104_130 |
0.0736969454 |
- |
- |
C-14 sterol reductase, putative [Ricinus communis] |
| 3 |
Hb_000107_110 |
0.0760591332 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 4 |
Hb_002603_050 |
0.0771166948 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP20-1-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_003052_180 |
0.091915957 |
- |
- |
Diacylglycerol Cholinephosphotransferase [Ricinus communis] |
| 6 |
Hb_033152_070 |
0.0961595127 |
- |
- |
N-acetyl-gamma-glutamyl-phosphate reductase, putative [Ricinus communis] |
| 7 |
Hb_000928_190 |
0.1043756503 |
- |
- |
PREDICTED: putative acyl-activating enzyme 19 isoform X1 [Jatropha curcas] |
| 8 |
Hb_003207_020 |
0.1059072687 |
- |
- |
PREDICTED: bifunctional dTDP-4-dehydrorhamnose 3,5-epimerase/dTDP-4-dehydrorhamnose reductase [Jatropha curcas] |
| 9 |
Hb_010042_020 |
0.1077410176 |
- |
- |
UDP-n-acteylglucosamine pyrophosphorylase, putative [Ricinus communis] |
| 10 |
Hb_012678_020 |
0.1087954715 |
- |
- |
PREDICTED: outer envelope protein 61 isoform X1 [Jatropha curcas] |
| 11 |
Hb_009661_030 |
0.1097275766 |
- |
- |
PREDICTED: SPX and EXS domain-containing protein 1-like [Jatropha curcas] |
| 12 |
Hb_002902_130 |
0.1110134671 |
- |
- |
PREDICTED: molybdate-anion transporter [Jatropha curcas] |
| 13 |
Hb_005218_020 |
0.1133829237 |
- |
- |
Uncharacterized protein isoform 3 [Theobroma cacao] |
| 14 |
Hb_002481_060 |
0.1135953992 |
- |
- |
PREDICTED: uncharacterized protein LOC105640851 [Jatropha curcas] |
| 15 |
Hb_000136_100 |
0.1146565023 |
- |
- |
PREDICTED: syntaxin-61 [Jatropha curcas] |
| 16 |
Hb_000014_040 |
0.1153290793 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000258_240 |
0.1154154775 |
- |
- |
sur2 hydroxylase/desaturase, putative [Ricinus communis] |
| 18 |
Hb_000004_020 |
0.1156630804 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_004046_030 |
0.1156904044 |
- |
- |
PREDICTED: GPCR-type G protein 1 [Jatropha curcas] |
| 20 |
Hb_003529_030 |
0.1160467663 |
- |
- |
Non-imprinted in Prader-Willi/Angelman syndrome region protein, putative [Ricinus communis] |