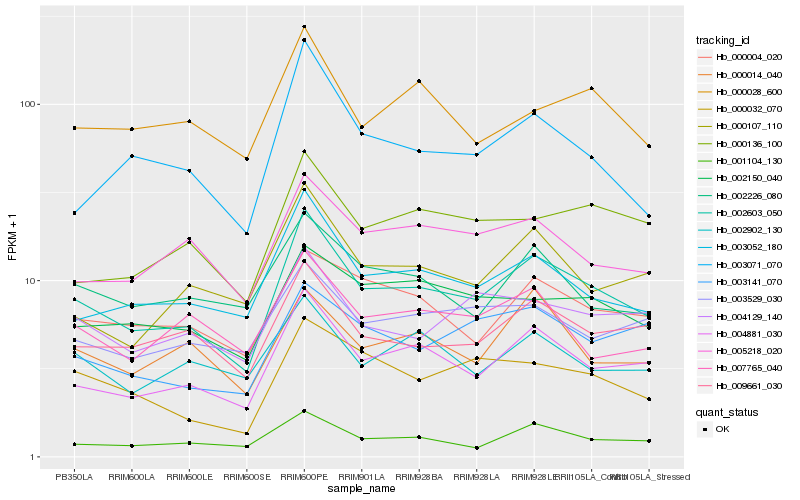
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002603_050 |
0.0 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP20-1-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_004881_030 |
0.0771166948 |
- |
- |
PREDICTED: uncharacterized protein LOC105629532 isoform X1 [Jatropha curcas] |
| 3 |
Hb_003052_180 |
0.0986631866 |
- |
- |
Diacylglycerol Cholinephosphotransferase [Ricinus communis] |
| 4 |
Hb_001104_130 |
0.1034630447 |
- |
- |
C-14 sterol reductase, putative [Ricinus communis] |
| 5 |
Hb_009661_030 |
0.1081440064 |
- |
- |
PREDICTED: SPX and EXS domain-containing protein 1-like [Jatropha curcas] |
| 6 |
Hb_000004_020 |
0.1179953301 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000032_070 |
0.118457694 |
- |
- |
hypothetical protein POPTR_0003s13760g [Populus trichocarpa] |
| 8 |
Hb_005218_020 |
0.118502465 |
- |
- |
Uncharacterized protein isoform 3 [Theobroma cacao] |
| 9 |
Hb_002150_040 |
0.1199073579 |
- |
- |
PREDICTED: CMP-sialic acid transporter 1 isoform X3 [Jatropha curcas] |
| 10 |
Hb_000107_110 |
0.1203069145 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 11 |
Hb_002226_080 |
0.1204258817 |
- |
- |
PREDICTED: uncharacterized protein LOC105641402 isoform X1 [Jatropha curcas] |
| 12 |
Hb_004129_140 |
0.121611691 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At1g06470 [Jatropha curcas] |
| 13 |
Hb_003071_070 |
0.1225948303 |
- |
- |
sucrose synthase 3 [Hevea brasiliensis] |
| 14 |
Hb_000014_040 |
0.1227096248 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000136_100 |
0.1228876305 |
- |
- |
PREDICTED: syntaxin-61 [Jatropha curcas] |
| 16 |
Hb_003529_030 |
0.1251568993 |
- |
- |
Non-imprinted in Prader-Willi/Angelman syndrome region protein, putative [Ricinus communis] |
| 17 |
Hb_007765_040 |
0.1252291244 |
- |
- |
Cytosolic enolase isoform 3 [Theobroma cacao] |
| 18 |
Hb_002902_130 |
0.1254958732 |
- |
- |
PREDICTED: molybdate-anion transporter [Jatropha curcas] |
| 19 |
Hb_003141_070 |
0.1277361874 |
- |
- |
PREDICTED: probable protein phosphatase 2C 51 [Jatropha curcas] |
| 20 |
Hb_000028_600 |
0.1281431356 |
- |
- |
glutaredoxin-1, grx1, putative [Ricinus communis] |