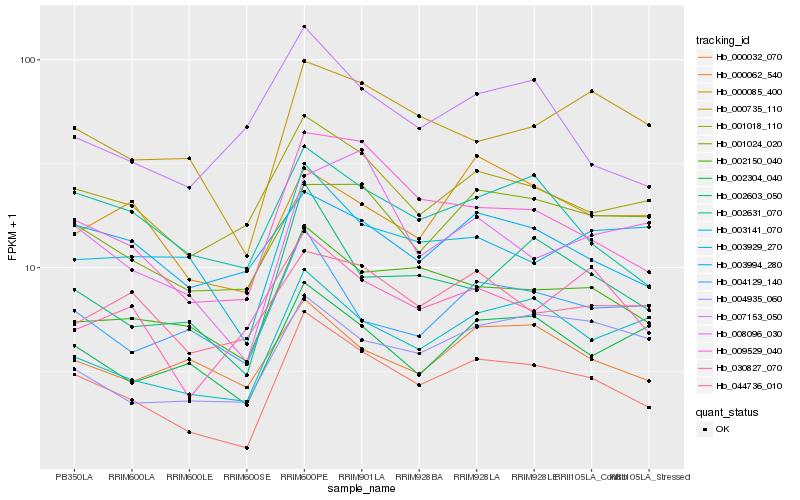
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000032_070 |
0.0 |
- |
- |
hypothetical protein POPTR_0003s13760g [Populus trichocarpa] |
| 2 |
Hb_009529_040 |
0.1062220023 |
- |
- |
PREDICTED: transmembrane protein 120 homolog isoform X1 [Malus domestica] |
| 3 |
Hb_002603_050 |
0.118457694 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP20-1-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_001018_110 |
0.1233140651 |
- |
- |
unnamed protein product [Coffea canephora] |
| 5 |
Hb_002150_040 |
0.1233854276 |
- |
- |
PREDICTED: CMP-sialic acid transporter 1 isoform X3 [Jatropha curcas] |
| 6 |
Hb_003141_070 |
0.1278802318 |
- |
- |
PREDICTED: probable protein phosphatase 2C 51 [Jatropha curcas] |
| 7 |
Hb_002631_070 |
0.129021416 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g16860-like [Jatropha curcas] |
| 8 |
Hb_000735_110 |
0.1376237277 |
- |
- |
fk506-binding protein, putative [Ricinus communis] |
| 9 |
Hb_001024_020 |
0.1380880521 |
- |
- |
hypothetical protein JCGZ_08989 [Jatropha curcas] |
| 10 |
Hb_002304_040 |
0.143261008 |
- |
- |
PREDICTED: uncharacterized protein LOC105649623 isoform X2 [Jatropha curcas] |
| 11 |
Hb_004129_140 |
0.1448654677 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At1g06470 [Jatropha curcas] |
| 12 |
Hb_030827_070 |
0.1466449032 |
- |
- |
PREDICTED: transmembrane protein 161B [Jatropha curcas] |
| 13 |
Hb_003994_280 |
0.1470600946 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g16860-like [Jatropha curcas] |
| 14 |
Hb_004935_060 |
0.1472994612 |
- |
- |
Uncharacterized protein isoform 2 [Theobroma cacao] |
| 15 |
Hb_003929_270 |
0.1498994294 |
- |
- |
S-adenosylmethionine-dependent methyltransferase, putative [Ricinus communis] |
| 16 |
Hb_000062_540 |
0.1509803683 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_007153_050 |
0.1524207613 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 8 [Jatropha curcas] |
| 18 |
Hb_044736_010 |
0.1527793351 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 19 |
Hb_008096_030 |
0.1540963879 |
- |
- |
PREDICTED: uncharacterized protein LOC105638438 [Jatropha curcas] |
| 20 |
Hb_000085_400 |
0.1549893642 |
- |
- |
Heterogeneous nuclear ribonucleoprotein 27C, putative [Ricinus communis] |