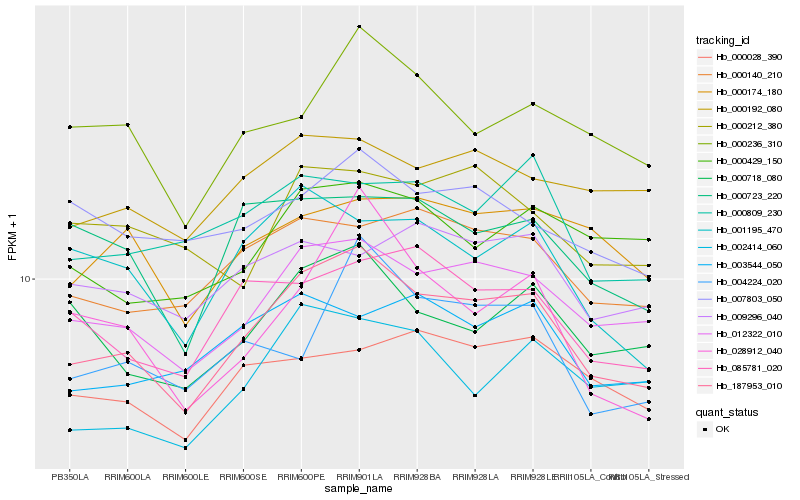
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_187953_010 |
0.0 |
- |
- |
PREDICTED: type II inositol 1,4,5-trisphosphate 5-phosphatase FRA3 isoform X2 [Jatropha curcas] |
| 2 |
Hb_012322_010 |
0.0701610835 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 3 |
Hb_000140_210 |
0.099700106 |
- |
- |
sec10, putative [Ricinus communis] |
| 4 |
Hb_000718_080 |
0.1012976493 |
- |
- |
hypothetical protein PRUPE_ppa005259mg [Prunus persica] |
| 5 |
Hb_085781_020 |
0.1054774428 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 1 [Jatropha curcas] |
| 6 |
Hb_000236_310 |
0.1078789619 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1-like [Malus domestica] |
| 7 |
Hb_001195_470 |
0.1089850605 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g75140 [Jatropha curcas] |
| 8 |
Hb_000809_230 |
0.1113162938 |
- |
- |
PREDICTED: uncharacterized protein LOC105639681 isoform X1 [Jatropha curcas] |
| 9 |
Hb_002414_060 |
0.1118386462 |
- |
- |
hypothetical protein EUGRSUZ_E01310 [Eucalyptus grandis] |
| 10 |
Hb_000723_220 |
0.1121977452 |
- |
- |
PREDICTED: uncharacterized protein LOC105642234 [Jatropha curcas] |
| 11 |
Hb_003544_050 |
0.1129338852 |
- |
- |
PREDICTED: uncharacterized protein LOC105644300 isoform X1 [Jatropha curcas] |
| 12 |
Hb_004224_020 |
0.1138796861 |
- |
- |
PREDICTED: uncharacterized protein LOC105643786 [Jatropha curcas] |
| 13 |
Hb_000192_080 |
0.1149256941 |
- |
- |
PREDICTED: serine/threonine-protein kinase SAPK3 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000174_180 |
0.1149798277 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 15 |
Hb_000028_390 |
0.1151924557 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 3-like [Jatropha curcas] |
| 16 |
Hb_000429_150 |
0.1181550172 |
- |
- |
hypothetical protein JCGZ_06799 [Jatropha curcas] |
| 17 |
Hb_000212_380 |
0.1184126408 |
- |
- |
Aspartic proteinase Asp1 precursor, putative [Ricinus communis] |
| 18 |
Hb_009296_040 |
0.1189347374 |
- |
- |
hypothetical protein L484_007435 [Morus notabilis] |
| 19 |
Hb_007803_050 |
0.1189678064 |
- |
- |
PREDICTED: uncharacterized protein LOC105129170 isoform X2 [Populus euphratica] |
| 20 |
Hb_028912_040 |
0.1194872107 |
- |
- |
hypothetical protein POPTR_0016s03250g [Populus trichocarpa] |